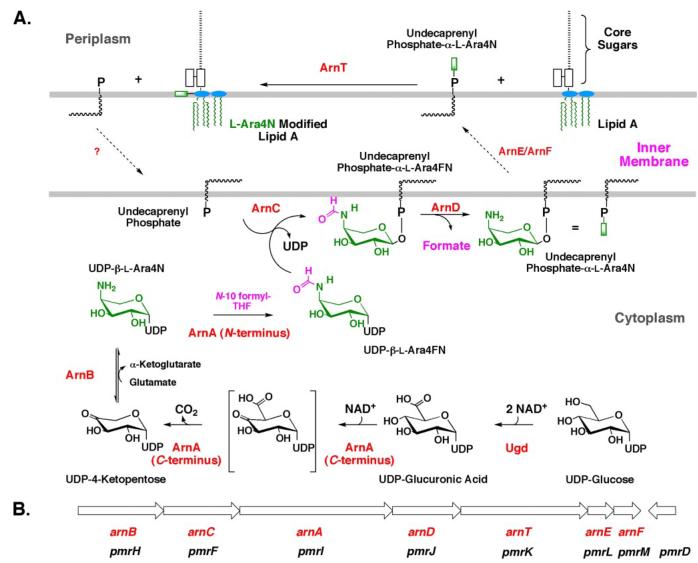
FIGURE 2. Biosynthesis of undecaprenyl phosphate-α-L-Ara4N and transfer of the L-Ara4N moiety to lipid A.
A, in polymyxin-resistant E. coli and S. typhimurium, biosynthesis of the L-Ara4N moiety begins with oxidation of UDP-glucose to UDP-glucuronic acid (2). Next, the C-terminal domain of ArnA catalyzes the NAD+-dependent oxidative decarboxylation of UDP-glucuronic acid to yield an unusual UDP-4-ketopentose (30, 32), which is converted by the transaminase ArnB to UDP-β-L-Ara4N (29). Subsequently, the N-terminal domain of ArnA uses N-10-formyltetrahydrofolate to N-formylate UDP-β-L-Ara4N (28, 30, 32). ArnC (a distant orthologue of dolichyl phosphate-mannose synthase) selectively transfers the L-Ara4-formyL-N residue to undecaprenyl phosphate (28). Next, ArnD catalyzes deformylation of this substance to undecaprenyl phosphate-α-L-Ara4N (13), preventing the reversal of the ArnC-catalyzed reaction (28). After transport to the outer surface of the inner membrane, presumably by ArnE (PmrL) and ArnF (PmrM) as shown in the present study, the membrane enzyme ArnT (31) transfers the L-Ara4N moiety to lipid A. B, order of genes and direction of transcription (left to right) of the pmr operon (16). The preferred arn terminology (2, 45), which is consistent with the generalized bacterial polysaccharide gene nomenclature (94), is shown in red. We suggest that the older pmr nomenclature be retained for the regulatory genes pmrA, pmrB, and pmrD, because their products have many other functions besides regulating the expression of enzymes needed for L-Ara4N biosynthesis.