Fig. 5.
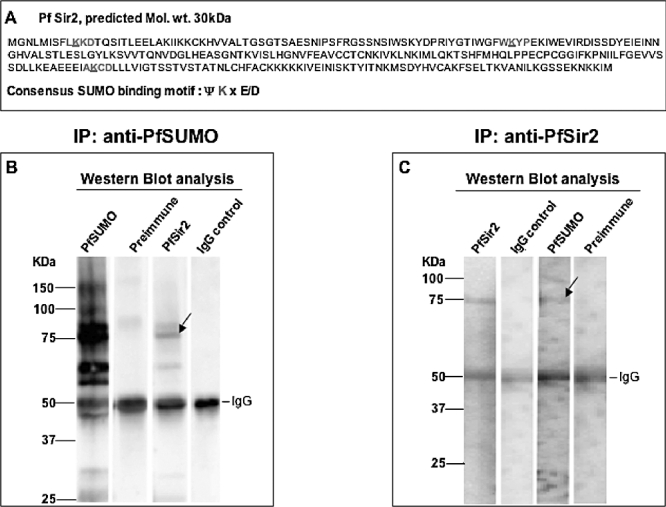
Immunoblot analysis of immunoprecipitated PfSir2 and sumoylated proteins.
A. Sequence of PfSir2 obtained from PlasmoDB tested on SUMOplot to scan for potential SUMO acceptor motifs (ψ K x E/D, with ψ – a hydrophobic residue, K – acceptor lysine to which the di-glycine motif of the SUMO peptide is attached, x – any amino acid followed by a Glutamic acid or Aspartic acid). Coloured amino acids depict probable motifs with red depicting high probability motif and blue lower probability, critical lysine residue with the motif is underlined.
B. Proteins immunoprecipitated from nuclear extracts with anti-PfSUMO sera were run on a 4–12% gradient gel and immunoblotted with anti-PfSUMO sera and pre-immune sera as positive controls (first two lanes), anti-PfSir2 (third lane) and anti-IgG (fourth lane). Arrow marks corresponding PfSir2 band(s) observed in anti-PfSUMO immunoprecipitated protein extract.
C. Proteins immunoprecipitated with anti-PfSir2 sera were run on a 4–12% gradient gel and immunoblotted with anti-PfSir2 sera and anti-IgG as positive controls (first two lanes), anti-PfSUMO (third lane) and anti-PfSUMO pre-immune sera (fourth lane). Arrow marks corresponding sumoylated fraction observed in anti-PfSir2 immunoprecipitated protein(s).
