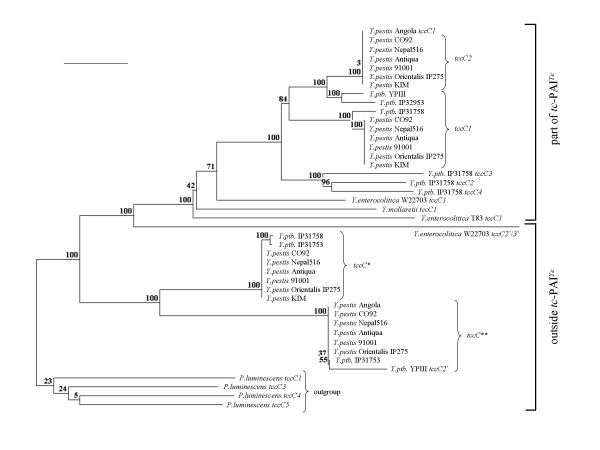
Figure 2.
Cladogram based on 39 TccC amino acid sequence data. Four TccC sequences of P. luminescens subspecies laumondii strain TT01 served as outgroup. The phylogenetic analysis was performed with the neighbour-joining method and calculated using the two parameter model of Kimura [31]. Values on each branch indicate the occurrence (%) of the branching order in 500 bootstrapped trees. *insertion site between genes coding for the insertion element IS1541 and a hypothetical protein, **insertion site between genes encoding a N-ethylmaleimide reductase and a lactoylglutathione lyase. The frameshift of tccC in strain W22703 was not considered here; hence, one coherent amino acid sequence was used for the alignment. Bar represents 0.1% sequence divergence. Y. ptb., Y. pseudotuberculosis.