Figure 5.
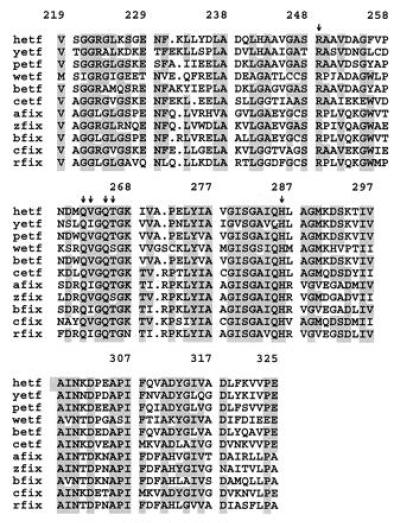
Sequence alignment of residues involved in binding FAD among members of the ETF α subunit family of proteins. Proteins used in the sequence alignment are: hetf, human ETF (32); petf, P. denitrificans ETF (33); yetf, Saccharomyces cerevisiae hypothetical ETF α subunit; wetf, Methylophilus methylotrophus W3A1 ETF (9); betf, Bradyrhizobium japonicum putative ETF (GenBank accession no. U32230U32230); cetf, Clostridium acetobutylicum putative ETF (34); afix, Azorhizobium caulinodans putative FixB (35); zfix, Azotobacter vinelandii putative FixB, (Protein Identification Resource accession no. S49188S49188); bfix, B. japonicum putative FixB (36); cfix, C. acetobutylicum putative FixB (GenBank accession no. M91817M91817); and rfix, Rhizobium meliloti putative FixB (37). Residues that are identical in at least six of the sequences are shaded. Arrows indicate residues involved in binding the isoalloxazine portion of the FAD, as shown in Fig. 4.
