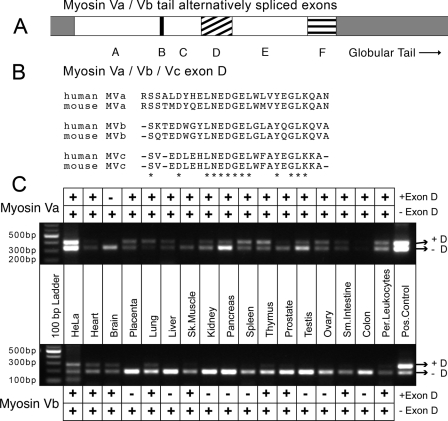
FIGURE 1.
Tissue distribution of human myosin Va and myosin Vb splice isoforms. A, schematic of the alternative exon organization in the tails of myosin Va and myosin Vb. It is known that exons B, D, and F are subject to alternative splicing in myosin Va, whereas there is only evidence that exon D is alternatively spliced in myosin Vb, which does not contain exon F. B, alignment of exon D sequences from mouse and human myosin V's. myosin Va and myosin Vb both contain exon D (amino acids 1320-1346 of myosin Va and 1315-1340 of myosin Vb), whereas myosin Vc contains an exon D-like region (amino acids 1124-1147 of human myosin Vc) that is not known to be alternatively spliced. Alignment of the exon D regions from all three motors reveals a high degree of homology, especially in the center of the exon. Asterisks indicate amino acid identities. C, PCR-based analysis of human tissue panels reveals the alternative splicing pattern of exon D in myosin Va and myosin Vb. Primers flanking the region encoding exon D for both motors were used to amplify cDNA from human MTC™ panels (Clontech). cDNA amplified from HeLa cell RNA as well as myosin Va and myosin Vb tail constructs were used as positive controls. Variants expressing exon D (upper bands) and lacking exon D (lower bands) were visible. Per., peripheral; Pos., positive.