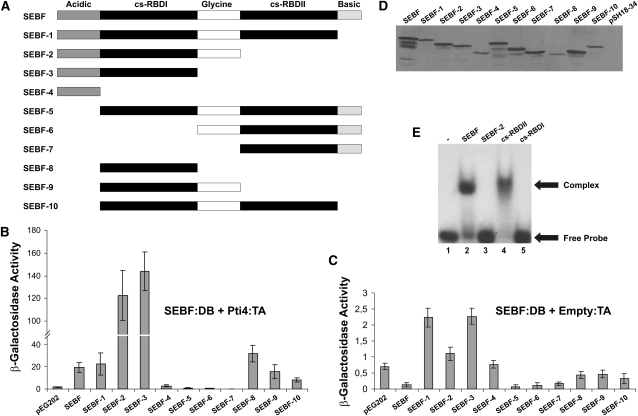
Figure 2.
SEBF Interacts with DNA through cs-RBDII and Recruits Pti4 via cs-RBDI.
(A) Schematics of SEBF and its deletion mutants analyzed in (B) to (E). Acidic and Basic indicate domains with these properties, while Glycine represents a Gly-rich region of SEBF. cs-RBDI and cs-RBDII refer to cs-RBDs I and II.
(B) Bar graph of β-galactosidase activity as the output of yeast two-hybrid assays testing the interaction of SEBF and the mutants depicted in (A) fused to the Lex-A DB domain with the full-length Pti4 fused to the B42 TA domain.
(C) Bar graph illustrating the background level of activity observed with each of the SEBF constructs depicted in (A) and coexpressed with the empty B42 TA vector.
For (B) and (C), results obtained by expressing the Lex-A DB domain alone (pEG202) along with Pti4:TA domain are shown as reference baseline values. Note the difference in scales between these panels. Values consist of n = 6 samples and represent averages ± 1 sd. Every bar represents an assay on three different colonies repeated on two independent transformation events.
(D) Immunoblot using an anti-SEBF antibody was performed to confirm the expression of SEBF and its mutant derivatives in cell lines used in (B).
(E) EMSA analyses were performed with the full-length SEBF (lane 2), SEBF-2 (lane 3), the cs-RBDI domain of SEBF (SEBF-8 in [A]; lane 4), and the cs-RBDII domain of SEBF (lane 5). All studies were done with 10 ng of purified recombinant protein and the 32P-labeled single-stranded SE oligonucleotide. Lane 1 contained only the labeled SE oligonucleotide.