Figure 5.
The SEBF-Pti4 Complex Represses Transcription in Vivo.
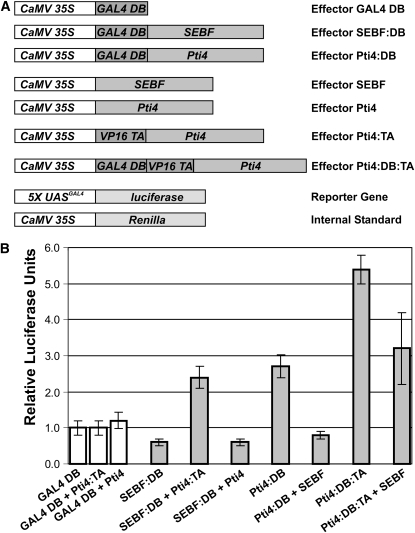
(A) Representations of the different constructs used in the plant two-hybrid and in vivo transcription assays. Promoters are shown in white boxes. CaMV 35S indicates the double cauliflower mosaic virus 35S:alfalfa mosaic virus promoter. 5X UASGAL4 indicates a promoter composed of a multimerized (five elements) Gal4 upstream activating sequence fused to a minimal TATA box and the Ω translational enhancer from the Tobacco mosaic virus. Coding sequences are shown in dark and light gray boxes. GAL4 DB indicates the GAL4 DNA binding domain. VP16 TA indicates the constitutive transactivation domain of viral protein 16. All constructs possess the polyadenylation signal from the nopaline synthase gene (data not shown). The 35S:Renilla construct is an internal reference to normalize transfection efficiency.
(B) Bar graph illustrating the interaction of Pti4 with SEBF as well as the transcriptional activation potential of Pti4, SEBF, and the SEBF-Pti4 complex. Each effector or pair of effectors was cotransfected with the reporter gene and the internal standard. The effector construct containing the GAL4 DB domain only was transfected into untreated leaves along with the reporter and internal standard constructs and was given an arbitrary value of 1 relative luciferase unit ± 1 sd after normalization with Renilla activity. All values are relative to the activity of Gal4 DB domain obtained in untreated leaves. Values consist of n = 25 samples and represent averages ± 1 sd. Every bar represents five bombardments repeated five times (n = 25). White bars represent Gal4 DB controls, while gray bars refer to data obtained with the proteins under investigation.