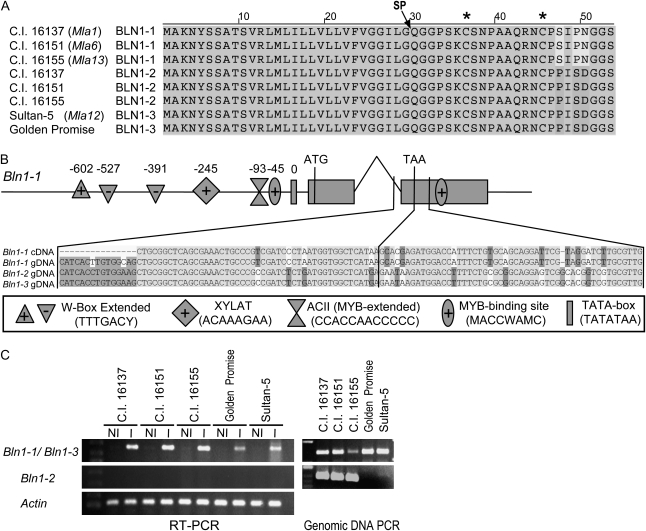
Figure 3.
Comparison of Bln1 sequences and transcript accumulation. A, Amino acid alignment of Bln1-1, Bln1-2, and Bln1-3 alleles/paralogs. Gray shaded boxes indicate identity across all eight sequences. SP designates the putative position of the signal peptide cleavage site, and asterisks at top designate the positions of the Cys residues. B, Bln1-1 gene model with promoter analysis of a genomic DNA fragment cloned via inverse PCR. Gray boxes indicate exons, with a single intron between exon 1 and 2. Symbols shown in the box at bottom indicate the positions of several promoter elements associated with defense (W-box, WRKY, MYB, P-box) and xylem- and root-specific expression. Alignment of genomic DNA of Bln1-1 and Bln1-2 and cDNA of Bln1-1 shows the extensive nucleotide divergence between these paralogs, beginning near the end of the intron. C, Differential transcript accumulation of Bln1 paralogs upon inoculation with Bgh isolate 5874. RT-PCR was performed on RNA isolated from seedling leaves 24 h after Bgh inoculation (I) or from noninoculated controls (NI). Actin was used as the internal control in all samples. The genomic DNA PCR results shown demonstrate the existence of different paralogs in different genotypes.