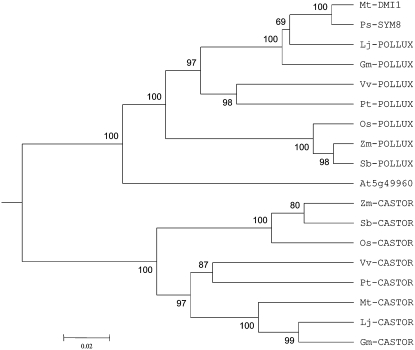
Figure 1.
Phylogenetic tree (unrooted) of CASTOR and POLLUX homologs in M. truncatula (Mt), L. japonicus (Lj), soybean (Gm), poplar (Pt), grapevine (Vv), Arabidopsis (At), rice (Os), sorghum (Sb), and maize (Zm). The tree was based on the C-terminal approximately 650 amino acids of the proteins. Sequence alignments were performed using ClustalX (Thompson et al., 1997) and manually curated. The tree was constructed by MEGA3.1 (Kumar et al., 2004), using the UPGMA method. Numbers below the branches represent the percentages of 1,000 bootstrap replications supporting the particular nodes.