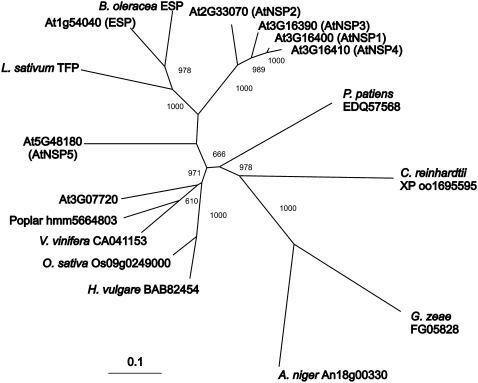
Figure 9.
Phylogeny of the Kelch domain. For all higher plant ESP homologues as well as a bryophyte, an algal, and two fungal homologues, the portion of each protein sequence containing the Kelch domains was used to generate a phylogeny. The neighbor-joining tree is shown with bootstrap values over 600 out of 1,000. A distance scale is included at the bottom for the protein tree. An unrooted tree is presented to allow a focus on relatedness. Source organisms were Arabidopsis, Populus trichocarpa, Vitis vinifera, Oryza sativa, Hordeum vulgare, Aspergillus niger, Giberella zeae, Chlamydomonas reinhardtii, Physcomitrella patiens, Lepidium sativum, and Brassica oleracea.