Table 1.
Activities of the 1,4-dihydropyridine derivatives as enhancers at the TRPV1 receptor ectopically expressed in NIH3T3 cells.
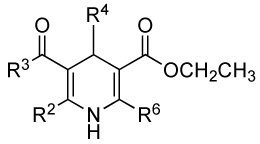 | ||||||||
|---|---|---|---|---|---|---|---|---|
| No | R2 | R3 | R4 | R6 | EC50(µM) ± SEM |
Hill Slope ± SEM |
Emax(%)a ± SEM |
nb |
| 3-Esters | ||||||||
| 7d | CH3 | CH3CH2O | CH3CH2 | Ph | — c | — | 383 ± 39 | 3 |
| 8d | CH3 | CH3CH2O | (CH3O) 2CH | Ph | — | — | 114 ± 24 | 2 |
| 9 | CH3 | CH3CH2O | CH3CH2 | 3-Cl-Ph | 17.1 ± 1.2 | 1.45 ± 0.28 | 347 ± 24 | 3 |
| 10 | CH3 | CH3CH2O | CH3CH2 | 3-F-Ph | 26.7 ± 1.1 | 1.98 ± 0.30 | 403 ± 21 | 3 |
| 11 | CH3 | CH3CH2O | CH3CH2 | 4-F-Ph | 29.2 ± 1.2 | 1.86 ± 0.42 | 372 ± 33 | 3 |
| 3-Thioesters | ||||||||
| 12d | CH3 | CH3CH2S | CH3CH2 | Ph | 28.1 ± 1.0 | 1.55 ± 0.17 | 435 ± 11 | 3 |
| 13 | CH3 | CH3CH2S | CH3CH2 | 2-F-Ph | — | — | 198 ± 2 | 2 |
| 14e | CH3 | CH3CH2S | CH3CH2 | 3-F-Ph | 21.8 ± 1.2 | 1.59 ± 0.33 | 446 ± 39 | 3 |
| 15 | CH3 | CH3CH2S | CH3CH2 | 4-F-Ph | 28.4 ± 1.2 | 1.82 ± 0.33 | 409 ± 32 | 3 |
| 16 | CH3 | CH3CH2S | CH3CH2 | 4-CN-Ph | — | — | 129 ± 2 | 2 |
| 17 | CH3 | CH3CH2S | CH3CH2 | cyclohexyl | — | — | 87 ± 8 | 2 |
| 18d | CH3 | CH3CH2S | CH3CH2CH2 | Ph | 22.4 ± 1.2 | 1.38 ± 0.23 | 364 ± 16 | 3 |
| 19 | CH3 | CH3CH2S | CH3CH2CH2 | 3-Cl-Ph | — | — | 177 ± 19 | 2 |
| 20 | CH3 | CH3CH2S | cPr | 4-F-Ph | — | — | 175 ± 4 | 2 |
| 21d | CH3CH2 | CH3CH2S | CH3CH2 | Ph | — | — | 257 ± 17 | 3 |
| 22d | CH3CH2CH2 | CH3CH2S | CH3CH2 | Ph | — | — | 148 ± 3 | 2 |
| 23d,e | CH3 | CH3OCH2CH2S | CH3CH2 | Ph | 21.3 ± 1.1 | 1.79 ± 0.17 | 626 ± 25 | 3 |
| 24 | CH3 | (CH3)2CHS | CH3CH2 | 2-F-Ph | — | — | 124 ± 4 | 2 |
| 25 | CH3 | PhCH2S | CH3CH2 | 2-F-Ph | — | — | 123 ± 1 | 2 |
| 26 | CH3 | PhCH2CH2S | CH3CH2CH2 | Ph | 27.6 ± 1.5 | 1.14 ± 0.32 | 360 ± 35 | 3 |
| 27 | CH3 | PhCH2CH2S | CH3CH2 | 2-F-Ph | — | — | 117 ± 4 | 2 |
| 28 | CH3 | PhCH2CH2S | CH3CH2 | 3-Cl-Ph | — | — | 194 ± 29 | 2 |
| 29 | CH3 | PhCH2CH2S | CH3CH2 | 4-F-Ph | 51.6 ± 1.6 | 1.18 ± 0.25 | 232 ± 7 | 3 |
| 30 | CH3 | PhCH2CH2S | CH3CH2 | 4-NO2-Ph | — | — | 101 ± 3 | 2 |
| 31 | CH3 | PhCH2S | CH3CH2 | 4-NO2-Ph | — | — | 98 ± 3 | 2 |
| 32 | CH3 | CH3CH2S | CH3CH2 | 2-naphthyl | — | — | 116 ± 19 | 2 |
Emax = TRPV1 activation at [Enhancer]max (100 µM), expressed as % of control (2 µM capsaicin).
“n” represents the number of separate assay plate runs, with compounds in duplicate wells per assay.
For compounds where EC50 and Hill Slope could not be calculated (i.e., the data could not be curve fitted), only the Emax is shown.
Ki value in binding to the human A3 adenosine receptor, in µM (corresponding compound number in ref. 27): 7, 2.27 (9); 8, 15.3 (14); 12, 2.01 (10); 18, 2.17 (12); 21, 0.907 (21); 22, 2.09 (22); 23, 4.58 (11).
Compound 23 is MRS1477; 14 is MRS3625.
