Abstract
IFT172, also known as Selective Lim-domain Binding protein (SLB), is a component of the Intraflagellar Transport (IFT) complex. In order to evaluate the biological role of the Ift172 gene, we generated a loss-of-function mutation in the mouse. The resulting Slb mutant embryos die between E12.5–13.0, and exhibit severe cranio-facial malformations, failure to close the cranial neural tube, holoprosencephaly, heart edema and extensive hemorrhages. Cilia outgrowth in cells of the neuroepithelium is initiated but the axonemes are severely truncated and do not contain visible microtubules. Morphological and molecular analyses revealed a global brain-patterning defect along the dorsal-ventral (DV) and anterior-posterior (AP) axes. We demonstrate that Ift172 gene function is required for early regulation of Fgf8 at the midbrain-hindbrain boundary and maintenance of the isthmic organizer. In addition, Ift172 is required for proper function of the embryonic node, the early embryonic organizer and for formation of the head organizing center (the anterior mesendoderm, or AME). We propose a model suggesting that forebrain and mid-hindbrain growth and AP patterning depends on the early function of Ift172 at gastrulation. Our data suggest that the formation and function of the node and AME in the mouse embryo relies on an indispensable role of Ift172 in cilia morphogenesis and cilia-mediated signaling.
Keywords: IFT172, Slb, cilia, node, nodal, MHB boundary
INTRODUCTION
The protein encoded by the mouse Ift172 gene was detected via its in vitro interaction with certain LIM-homeodomain transcription factors (Howard and Maurer, 2000). As a homolog of the IFT172 of the Chlamydomonas reinhardii, it belongs to a group of IFT (Intraflagellar Transport) proteins which are part of a multimeric protein complex that operates in cilia where it is transported along microtubules between the cell body and the tip of these external cell organelles. Mutations that affect components of the IFT machinery are known to compromise the formation and function of cilia (reviewed in Rosenbaum and Witman, 2002; Scholey and Anderson, 2006). The IFT172 protein is well conserved among different organisms (Pedersen et al., 2005). The IFT172 homolog Osm-1 of C. elegans is required for signal transduction of certain sensory neurons. Mutations in this gene affect the ability of the nematode to avoid high osmolarity (Perkins et al., 1986). In zebrafish, Ift172 has been shown to play an essential role in kidney cilia formation (Sun et al., 2004).
Numerous human diseases and congenital syndromes have been associated with cilia deficiencies or malfunctions, including polycystic kidney disease, situs inversus, retinal degeneration, and polydactyly (Bisgrove and Yost, 2006; Kulaga et al., 2004; Pan et al., 2005). Cilia play essential roles in the differentiation and survival of olfactory and retinal neurons and auditory hair cells (Tsujikawa and Malicki, 2004). Recent studies have uncovered an important role of vertebrate cilia components in cell-cell communication, including Shh (Corbit et al., 2005; Haycraft et al., 2005; Huangfu and Anderson, 2005; May et al., 2005; Huangfu et al., 2003; Huangfu and Anderson, 2006; Rohatgi et al., 2007), non-canonical and canonical Wnt (Jones et al., 2008; Park et al., 2006; Ross et al., 2005) and Platelet-Derived Growth Factor mediated signaling events (Schneider et al., 2005). These findings provide a link between ciliogenesis and known pathways of developmental regulation.
To evaluate the role of Ift172 in mouse development, we generated a loss-of-function mutation. The mutant phenotypes include defects in cilia morphogenesis, global defects in brain patterning, open neural tube defects and exencephaly. Another mutant allele of this gene, called wimple, was recently isolated via an ENU mutagenesis screen (Huangfu et al., 2003). The wimple mutant carries a point mutation that causes one amino acid substitution at the C-terminus of the IFT172 protein. The study of this mutant focused primarily on the observed ventral neural tube defect. In the study presented here we show that the Ift172 gene plays a crucial role in forebrain growth and patterning and in the maintenance of the isthmic organizer, a signaling center essential for the anterior-posterior patterning of the mid- and hindbrain regions. In addition, we find that Ift172 is required for proper function of the embryonic node, an early embryonic organizer, and for formation of the head organizing center, known as anterior mesendoderm (AME). We propose a model suggesting that the mechanism of global brain patterning depends on an early function of Ift172 at gastrulation and is linked to the role of this gene in cilia morphogenesis.
MATERIALS AND METHODS
Gene targeting and the generation of mutant mice
1.9 kb EcoRI and 2.5 kb XbaI/EcoRI fragments of the Ift172 gene were inserted into the Ostdupdel TKneo targeting vector (O. Smithies, University of North Carolina). The neo cassette present in the construct is inverted with respect to the direction of Ift172 transcription. The targeting construct was electroporated into R1 embryonic stem (ES) cells. DNA from ES cells was analyzed by PCR and Southern hybridization using a 3′ external probe (EcoRI-XbaI fragment). ES cells from two independent clones demonstrating the correct targeting event were injected into C57/BL-6 blastocysts. The Slb mutant allele was maintained in a 129/Sv x C57/BL-6 background by intercrossing. The genotyping of mice and embryos was performed by PCR. The mutant allele in heterozygous mice was identified using the following primers: Left: 5′-AGTTGAGCCTTCTTACGGGAAT-3′; right: 5′-TTGGCTACCCGTG ATATTGC-3′. To discriminate between the wild type and mutant alleles the following primer set was used: left: 5′CTCTCCCGATTCCACAAAAA-3′ ; right: 5′AGCCCTGGAGCTACATCAAA-3′.
RNA isolation and RT-PCR Analysis
Total RNA was isolated from embryos using the TRI reagent from Invitrogen, CA. RNA quantification was performed using RiboGreen RNA Quantification reagent (Molecular Probes, Eugene, Oregon). For RT-PCR we used the Cloned AMV first-strand synthesis kit from Invitrogen. The sequences of Ift172 primers used for RT-PCR analysis are as follows: Left primer: 5′-AAAGTTCGCGACTCAGGAAG-3′; right primer: 5′-TGCTTCTTTGACAAGGTCCA-3′. The Glycerol Aldehyde Phosphate Dehydrogenase (GAPDH) control primer-set was used as a positive control.
Whole-mount in situ hybridization, TUNEL assay and histology
Whole-mount in situ hybridization with digoxigenin-labeled RNA probes was performed essentially as described by Wilkinson et al., 1989. Modifications of the protocol include: fixing E7.0–7.5 embryos in 4% paraformaldehyde for 3 hrs, washing in PBS (pH7.4), bleaching with 6% hydrogen peroxide for 30 min and treating with 15μg/ml of proteinase K (Roche, IL) for 3 min. Embryos were otherwise processed following the standard protocol. The hybridization signals were detected with alkaline phosphatase-conjugated anti-digoxigenin antibodies and BM-purple substrate (Roche, IL).
The following probes were used in this study: Fgf8 (previously described in Gorivodsky et al., 2003), Wnt1 (AP. McMahon, Harvard University), Pax2 (A. Simeone, Medical Research Council, King’s College, UK), Otx2 (A. Simeone, Medical Research Council, King’s College, UK), Twist1 (Wolf et al., 1991), Krox-20 (DG. Wilkinson, National Institute for Medical Research, UK.), Kreisler (Cordes S., Mount Sinai Hospital, NY), Hoxa2 (R. Krumlauf, Stowers institute for Medical Research), Gbx2 (P. Chambon, Institut de Génétique et de Biologie Moléculaire et Cellulaire INSERM, France), Shh (AP. McMahon, Harvard University), Gli1 (A. L. Joyner, New York University, School of Medicine), Nodal (T. Yamaguchi, NCI, NIH), Foxa2 (S. L. Ang, MRC National Institute for Medical Research, UK) and Gsc (R. Behringer, University of Texas M. D. Anderson Cancer Center).
The fragment corresponding to nucleotides 1536–2212 of Ift172 cDNA (NM_026298) was isolated by PCR and cloned into the pBluescript SK cloning vector (Stratagene, USA) in order to generate the Ift172-specific probe.
The TUNEL assay was performed according to the manufacturer’s recommendations (Cell Death kit, Roche, IL). Whole-mount TUNEL reaction on E8.5 embryos was carried out as described previously (Chi et al., 2003). Anti-DIG antibody conjugated with Horse Radish Peroxidase enzyme (Roche, IL) and Fast-DAB substrate (Sigma-Aldrich, St. Louis, MO) was used to detect apoptotic cells. Embryos were cleared in glycerol and photographed.
For histology embryos were fixed in 4% paraformaldehyde, dehydrated, embedded in paraffin and sectioned. Sections were stained with hematoxylin and eosin.
Transmission Electron Microscopy
Fixation and embedding in plastic was performed according to electron microscopy standard protocols. Micrographs were taken on a Morgagni electron microscope (FEI company Eindhoven, Netherlands) with a MegaviewII CCD camera (Soft Imaging Systems) and AnalySis software (Olympus). The length of the primary cilia was determined by analyzing consecutive 200 nm sections for the presence of cilia in adjacent sections and once having identified cilia, which were present in their full extent within one section (and not obliquely cut), measuring the distance from the apical end of the basal body to the tip of the cilium.
RESULTS
The expression pattern of Ift172 mRNA
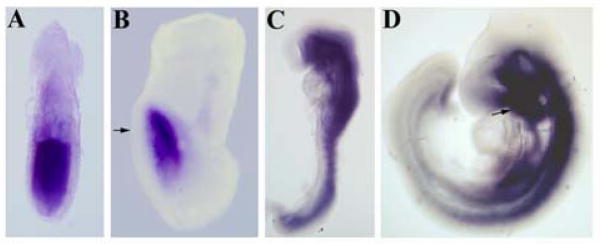
We analyzed the expression pattern of Ift172 mRNA at different stages of embryonic development. In early gastrulating embryos (E6.5), Ift172 is expressed throughout the whole epiblast, while at late primitive streak stages (E7.75–E8.0) transcripts are localized mainly in the anterior neuroectoderm (Fig. 1A and B). While the neuroectoderm is prominently stained, the signal is missing in the node and notochord. Ift172 would be expected to be expressed and to support cilia formation in these structures as well, albeit possibly at levels below our limit of detection. Alternatively, Ift172 transcription in the cells of the node may be downregulated, whereas IFT172 protein activity may persist.
Figure 1. Expression pattern of Slb mRNA.
(A) At E6.5 Ift172 is expressed throughout the epiblast. (B) At E7.75 Ift172 expression is restricted to anterior neuroectoderm. No signal is detected in the AVE (arrow). (C) At E8.5 Ift172 is strongly expressed in neural tube and somites. (D) At E9.5 Ift172 mRNA expression is localized to the ventral regions of the brain and neural tube. Strong expression of Ift172 is also detected in the branchial arches (arrow).
At E8.5, Ift172 is strongly expressed in the ventral neural tube and the somites and weakly in the dorsal neural tube (Fig. 1C). By E9.0–9.5, Ift172 transcripts become localized predominantly to the ventral part of the neural tube and the brain. We also observed strong signals in the first branchial arch, the outflow tract of the heart and the aortic arches (Fig. 1D). This pattern of Ift172 mRNA expression suggests that itsfunction is required for early embryonic patterning during gastrulation and later in the neural tube and the cardiovascular system.
Targeted deletion of the Ift172 gene and the mutant phenotype
To target the Ift172 gene, we inserted a neo cassette replacing the first three exons (Fig. S1A). We confirmed a correct targeting event in ES cells by Southern blot hybridization showing deletion of the 5′ end of the gene (Fig. S1B). We refer to the mutant allele as Slb. Reverse transcription-PCR analysis with primers corresponding to the deleted region detected a band of expected size in RNA isolated from wild type but not mutant embryos (Fig. S1C). The 3′ portion of the gene is still expressed in the mutant tissues (Fig. S1C). While we cannot exclude the possibility that this remaining portion of the gene gives rise to truncated IFT172 proteins, we can nevertheless rest assured that our mutants carry a hypomorphic allele since their phenotype closely resembles that of the previously published wimple mutant allele of Ift172 (Huangfu et al., 2003).
Slb mutant embryos are viable until E12.5. The mutant embryos show a neural tube defect (NTD) and holoprosencephaly with 87% penetrance (n=92). At E9.5, the cranial neural tube fails to close, leading to exencephaly by E12.5.
All mutant embryos demonstrate truncation of the telencephalon and diencephalon and lack eyes. However, the optic vesicle is visible at E9.5 suggesting that eye induction is not compromised. We also observed severe cranio-facial defects with a hypertrophic maxillar prominence of the first branchial arch (Fig. 2B and C).
Figure 2. Holoprosencephaly and brain growth and patterning defects in Slb mutant embryos.
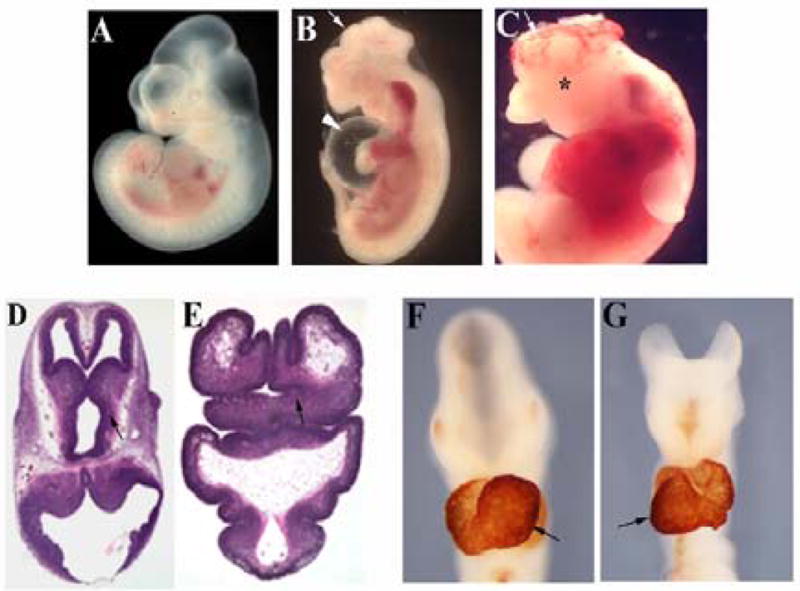
(A, B) Wild type and Slb mutant embryos at E11.5, respectively. (B) A representative Slb embryo showing severe craniofacial and brain malformations with exencephaly (arrow). Heart edema is clearly visible (arrowhead). (C) E11.75 Slb embryo. The cranial neural tube fails to close at the level of fore- and midbrain (arrow). The mutant embryo also displays severe hemorrhages. No eye formation is detected (asterisk). (D, E) H&E staining of brain sections collected from E12.5 wild type and Slb mutant embryos respectively. (E) The mutant brain shows disorganized forebrain structures with a severely reduced diencephalon (arrow), a phenotype characterized as lobar and/or semilobar holoprosencephaly. (F, G) These panels show E9.5 wild type and mutant embryos respectively. The hearts are stained with anti-cardiac Myosin (MF-20) antibody. In the Slb mutant embryo (G), heart looping is reversed compared to the wild type embryo (arrows in F and G).
We noted a randomization of left-right asymmetry in Slb mutant embryos. In 42% of observed mutant embryos (n=92) heart looping is inverted compared to the wild type heart (Fig. 2F and G). The outflow tract is remarkably shorter in the mutant, and the right ventricle is reduced in size. The mutation typically causes heart edema (Fig. 2B) and hemorrhages and leads to death by E13.0. Finally, about one in six mutant embryos analyzed at E12.5 show polydactyly suggesting defects in AP limb axis formation (data not shown).
Altered cilia morphology in Slb mutants
Since IFT172 is a component of the IFT complex whose assembly and function is critical for cilia morphogenesis, we performed a detailed ultrastructural study of the primary cilia in Slb mutant embryos. We analyzed cilia formation in embryonic neural tubes at E8.5 because at this stage Ift172 expression is abundant in the brain. Using transmission electron microscopy, we examined a series of ultrathin sections from various forebrain, midbrain and hindbrain regions, including the area around the midbrain-hindbrain boundary (MHB).
The cilia are located on the apical cell surface and have both extracellular and intracellular components. The extracellular portion is a cylindrical structure, 250–300 nm in diameter, which extends from the cell body. It consists of a specialized membrane and an inner core of microtubules and specific proteins called the axoneme. The axoneme is anchored to the basal body, which originates from the centriole (Hagiwara et al., 2000).
The structure of the basal body of the cilia of Slb mutant cells resembles that of wild type cells, suggesting that the intracellular portion of the Slb mutant cilium is not affected. Axonemes of neuroepithelial cilia from wild type embryos have a well-organized microtubule apparatus (Fig. 3A). In contrast, cilia from Slb mutant embryos possess very short axonemes with no visible microtubules (Fig. 3B, C, and D), although in rare cases a thin membranous sheath is present (Fig. 3B). We conclude that Ift172 gene function is not required for the induction of cilia per se, yet the truncated, abnormally shaped cilia formed by mutant cells are likely non-functional. Since these morphological changes were observed in a variety of brain sections, it is entirely possible that Ift172 gene function is essential for cilia morphogenesis in all Ift172-expressing cells of the developing brain.
Figure 3. Defect in cilia morphogenesis.
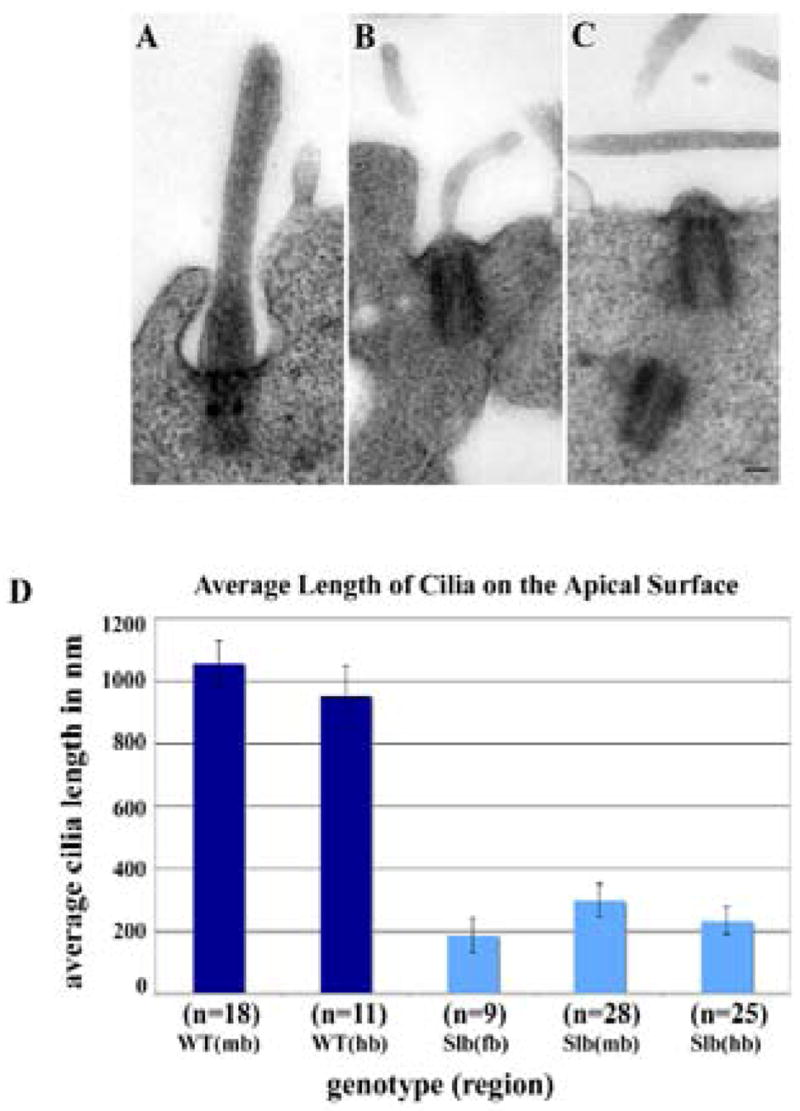
Primary cilia in the neuroepithelium of Slb mutants are greatly reduced in length. (A–C) Primary cilia from E8.5 wild type and Slb mutant neuroepithelium. Electron micrographs are taken from 200 nm sections through the mid/hindbrain area of wild type (A) and Slb mutant (B, C) embryos. The scale bar is 100 nm. (D) Average length of primary cilia on the apical surface of neuroepithelial cells in different brain regions. The average length of cilia was determined on 200 nm sections of different brain regions (fb: forebrain; mb: midbrain; hb: hindbrain) of one wild type (WT) and two Slb (SLB) E8.5 mouse embryos (n: number of cilia analyzed).
Defect in anterior hindbrain patterning
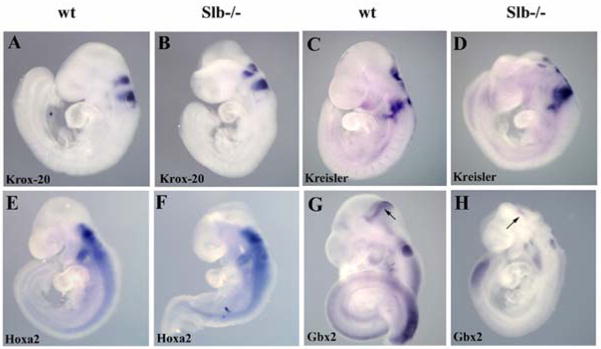
The shortcomings in cilia formation of Slb mutant brain cells suggests that signaling events mediated by the IFT machinery are adversely affected, resulting in the observed defects in brain patterning and development. In order to study Ift172 involvement in hindbrain development, we examined several transcription factors that show rhombomere-specific expression patterns. The hindbrain is composed of seven rhombomeres: r1–r7. The transcription factor Mafb (Kreisler) marks r5 and r6 and is critical for hindbrain segmentation posterior to r4 (Cordes and Barsh, 1994). Egr2 (Krox20) is specifically expressed in r3 and r5 (Wilkinson et al., 1989) and plays a crucial role in the maintenance of segment identity in the hindbrain (Schneider-Maunoury et al., 1993). Both are expressed normally in Slb mutant embryos (Fig. 4A–D). The Hoxa2 homeobox gene marks r2–r7 (Manley and Capecchi, 1995) and is required to specify the identity of the r2/r3 segment (Davenne et al., 1999; Gavalas et al., 1997). The expression pattern of Hoxa2 was similar for wild type and mutant embryos (Fig. 4E and F). In contrast, Gbx2 which is expressed in r1 and the posterior domain of the isthmic organizer (Liu et al., 1999), was dramatically reduced in the mutant brain at E9.5 (n=5; Fig. 4G and H), suggesting that r1 formation is adversely affected in the Slb mutant.
Figure 4. Expression analysis of hindbrain markers.
(A, B) At E9.5 Krox 20 mRNA expression in rhombomeres r3 and r5 is comparable in wild type (A) and Slb mutant (B) embryos. (C–F) No difference in the expression patterns of Kreisler and Hoxa2 mRNAs between wild type (C and E, respectively) and mutant embryos (D and F) was observed. (G, H) Gbx2 mRNA is greatly reduced in the mutant (arrow in H) compared to the wild type (arrow in G).
A role for Ift172 in the maintenance of the MHB boundary and forebrain development
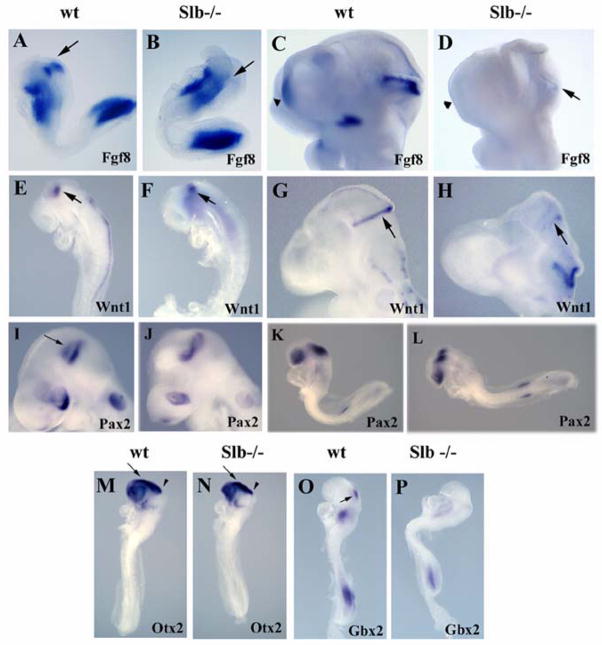
The isthmic organizer plays a crucial role in the development of the first rhombomere. We examined the formation and correct positioning of the organizer in Slb mutants by studying the expression of the genes which mark the region. The Fgf8 gene plays a major role in controlling the function of this organizer. In wild type embryos, Fgf8 transcripts begin to accumulate at the MHB boundary at the 3–4 somite stage (Crossley and Martin, 1995). Between E8.5 and E9.5 the Fgf8 expression pattern becomes more refined, and by E10.5–11.0 it appears as a sharp boundary.
In Slb mutant embryos (n=7), a severe reduction in Fgf8 mRNA expression levels at the MHB boundary is observed as early as E8.3 (Fig. 5A and B). At later stages, Fgf8 transcripts are strongly reduced in the isthmic region (Fig. 5C and D). Wnt1 activity is also critical in this context. Its expression precedes that of Fgf8 and is known to regulate Fgf8 expression (McMahon et al., 1992). In wild type embryos, Wnt1 expression has been observed at the MHB boundary and in the dorsal neural tube at E8.5–9.5 (McMahon and Bradley, 1990; McMahon et al., 1992). While Wnt1 is expressed at a normal level at the MHB boundary in E8.5 Slb mutants, it is lost in the dorsal neural tube (n=4; Fig. 5E and F). By E9.5, Wnt1 is also drastically reduced at the MHB boundary, and fails to be induced in the dorsal brain (n=5; Fig. 5G and H).
Figure 5. Defects in isthmic organizer formation and forebrain development.
(A, B) At E8.3, expression of Fgf8 at the MHB boundary (arrow) is severely downregulated in Slb mutant embryos (B) as compared to the wild type (A). (C, D) AtE9.5, Fgf8 is present at the MHB boundary of the wild type embryo (C), butis barely detectable in the Slb mutant (arrow in D). Fgf8 transcripts are normally expressed in the commissural plate (CP) of the wild type embryo, but are not found in the forebrain of the mutant (arrowheads in C and D). (E, F) At E8.5, Wnt1 expression at the MHB boundary (arrow) of the Slb mutant embryo (F) is comparable to that of the wild typeembryo (E), but missing in the dorsal neural tube. (G, H) At E9.5 the Wnt1 expression at theMHB boundary is maintained in the wild type (arrow in G), but is virtually lost in the Slb mutant embryo (H). (I, J) The most anterior Pax2 expression domain atthe MHB boundary (arrow in I) is absent in the Slb mutantembryo (J). (K, L) Pax2 expression is not reduced in the mutant at E8.5 (L) compared to the wild type embryo (K). (M, N) Otx2 expression in the forebrain (arrow) and midbrain (arrowhead) regions of E8.5 Slb mutant embryo (N) is comparable to that of the wild type embryo (M). (O, P) At E8.5, Gbx2 is severely affected in the Slb mutant (P) compared to the wild type (arrow in O).
Pax2, another gene prominently expressed in the isthmic organizer, is required to refine the domain of Fgf8 expression. Pax2 is normally expressed at the MHB boundary and in the optic vesicles (Schwarz et al., 1997). At the MHB boundary, itsexpression precedes and regulates that of Fgf8 (Ye et al., 2001). Once turned on, Fgf8 regulates Pax2 expression via a positive feedback loop (Schwarz et al., 1997). In E8.5 Slb mutant embryos, the Pax2 expression pattern resembles that of the wild type embryo (Fig. 5K and L). At E9.5 however, the anterior expression domain of Pax2 mRNA at the MHB boundary is absent in Slb mutants (n=4; Fig. 5I and J), most likely a consequence of diminished Fgf8 levels.
The anterior limit of the isthmic organizer is determined by the Otx2 expression domain (Broccoli et al 1999), while the posterior margin of the organizer is marked by Gbx2 expression (Liu et al., 1999). These two transcription factors negatively regulate each other, and deletion of one leads to an ectopic expansion of the other. Their function is required for proper positioning of the MHB boundary (Liu and Joyner, 2001). In the Slb mutant, Otx2 (n=5) is expressed normally at E8.5 in the fore- and midbrain regions, including the MHB boundary (Fig. 5M and N), while Gbx2 (n=6) expression is significantly reduced (Fig. 5O and P). Gbx2 expression in r1 is sufficient to repress Otx2 in the developing hindbrain (Liu and Joyner, 2001). Although a severe Gbx2 reduction is observed in the Slb mutant, there is no noticeable expansion of the Otx2 expression domain. Residual Gbx2 function in the Slb mutant might suffice to maintain the boundaries of Otx2 expression. This may serve to explain why the isthmic organizer is correctly positioned at the junction of the mid- and hindbrain of the mutant.
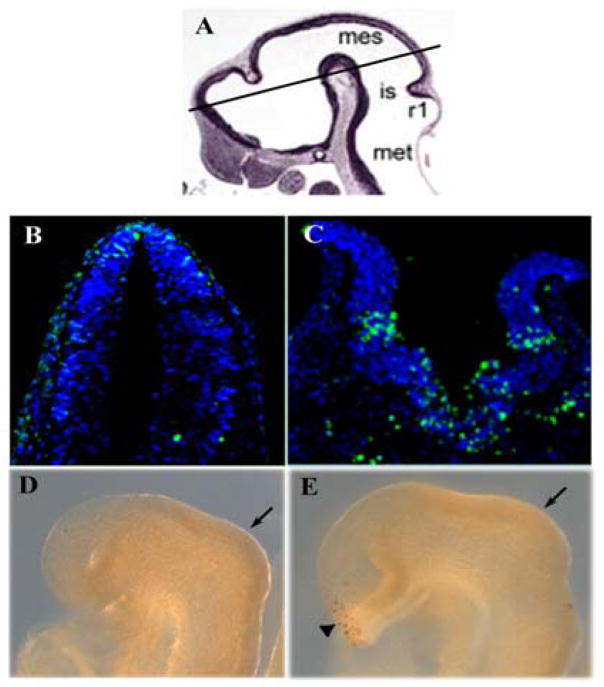
Failure to maintain the expression of the MHB boundary-specific signaling molecules and transcription factors may conceivably be due to increased cell death. Therefore, we analyzed the level of apoptosis of Slb mutant embryos at different stages of brain development. At E8.5 (9–10 somites) no increase in cell death is detected by TUNEL assay at the MHB boundary of the mutant embryo.
Expression of both Otx2 and Pax2 is similar in both wild type and mutant embryos at E8.5 (Fig. 5). By E9.5 signals for Pax2 are still found in the forebrain of the mutant. In the optic vesicle of the wild type embryo, Pax2 mRNA is localized in more distal ventral parts. In the mutant, Pax2 expression was reduced and ectopic signals were detected in the media-dorsal part of the optic vesicle (Fig. 5I and J). Expression of Fgf8 was attenuated in the anterior neural ridge at E8.5 (data not shown) and abolished in the commissural plate at E9.5 (Fig. 5C and D). Fgf8 deficiency is positively correlated with increased cell death in the forebrain region, as already seen at E8.5 (Fig. 6D and E). By E10.0 (23–25 somites), the level of cell death has increased dramatically in the mutant brain (Fig. 6B and C). These data suggest that Ift172 is required for maintenance of Fgf8 in the forebrain and for cell survival.
Figure 6. Cell death analysis.
Cell death in the wild type and Slb mutant embryos at E10.0 (B, C) and E8.5 (D, E), respectively. TUNEL assay reveals increased cell death in the brain of the Slb mutant embryo at E10.0 (C) compared to the wild type embryo (B). At E8.5 no increase in apoptosis is detected in the isthmic region (arrows) of the Slb mutant (E) as compared to the wild type embryo (D). Numerous apoptotic cells are detected in the ventral forebrain (arrowhead) of the mutant. (A) A histological section of E10.0 brain is used to show the plane of section in (B, C). Mes, Mesencephalon; is, isthmus; r1, rhombomere 1; met, metencephalon.
Collectively, our data indicate that expression patterns of Pax2, Wnt1 and Gbx2, the factors whose function is critical for isthmic organizer activity, are appropriately induced but fail to be maintained in Slb mutant embryos. We conclude that an early disruption of Fgf8 expression and subsequent increase of cell death in the isthmic region account for the defect in mid-hindbrain patterning.
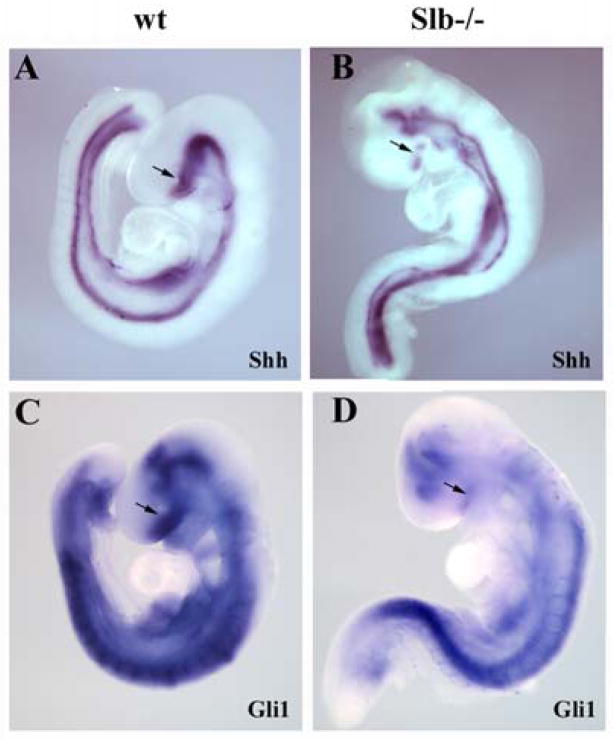
Studies of wimple and other IFT mutants have revealed an interaction between the intraflagellar transport mechanism and hedgehog (Hh) signaling (Huangfu et al., 2003; Huangfu and Anderson, 2005). In fact, several elements of the Hh pathway have been located to primary cilia (Corbit et al., 2005; Haycraft et al., 2005; Rohatgi et al., 2007). It therefore stands to reason that the defects in cilia formation that we detected in the Slb mutant may lead to a Hh deficiency. Indeed, some phenotypes, including holoprosencephaly and midline defects, suggest that Hh signaling may be compromised in the Slb mutant. Shh expression is practically abolished in the ventral forebrain and reduced in the midbrain (Fig. 7A and B). Expression of Gli1, a direct target of Shh, is severely downregulated in both fore- and midbrain (Fig. 7C and D). This suggests that Ift172 either directly or indirectly regulates Hh pathway activity in the embryonic brain.
Figure 7. Shh signaling in Slb mutants.
(A, B) At E9.0, Shh is expressed in the ventral forebrain and midbrain, ventral neural tube and notochord of the wild type embryo (A). In the Slb mutant (B), Shh expression is severely reduced in the ventral forebrain (arrows in A and B), but appears normal elsewhere. (C, D) At E9.0, Gli1 mRNA expression is abolished in the forebrain (arrows) and severely reduced in the midbrain of the mutant (D) as compared to a wild type littermate (C).
A requirement forIft172 in AME formation and the regulation of nodal in the node
The reduction in forebrain size and defects in mid- and hindbrain development observed in the mutant suggest an earlier role of the Ift172 gene in development, most likely during gastrulation. In fact, the defect in cilia morphogenesis at the node of the Wim mutant and the failure to establish a correct left-right axis in both Slb and Wim mutants (Huangfu et al., 2003) strongly suggest a function for Ift172 in the node organizer.
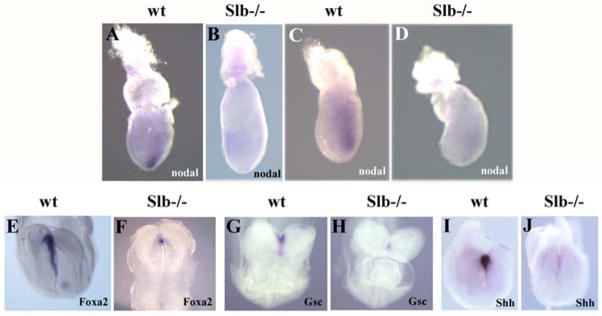
In an effort to determine the cause for the global defect of brain patterning in Slb mutant embryos, we analyzed the expression of nodal, a key molecular marker of the node. In early streak stage wild type embryos, nodal is expressed in the posterior epiblast. By the mid-streak stage, its transcripts are localized to the embryonic node (Conlon et al., 1994; Zhou et al., 1993). However, in the Slb mutant nodal expression is drastically reduced in the epiblast at E7.0 (Fig. 8C and D), as well as in the node at E7.5 (n=6; Fig. 8A and B). This strongly suggests that the function of the node in the Slb mutant is compromised. The observed role of Ift172 at gastrulation, prior to the onset of Shh expression thought to occur at E7.5, suggests an early, Shh-independent function that requires further study.
Figure 8. Expression of nodal and AME marker genes.
(A, B) At E7.5 nodal mRNA isexpressed in the node of a wild type embryo (arrow in A), but is barely detectable in a Slb mutant embryo (B). (C, D) At E7.0 nodal transcripts are localized in the posterior epiblast of the wild type embryo (C), while its expression is severely reduced in the Slb mutant embryo (D). (E, F) At E8.2, Foxa2 is expressed in the AME of the wild type embryo (E), but is severely downregulated in the Slb mutant (F). (G–J) Wild type expression of Gsc and Shh in the AME are shown in embryos at E8.5 (G) and E8.2 (I), but are strongly reduced in Slb mutant embryos (H and J), respectively.
The generation of the AME during late gastrulation stages is controlled by Nodal signaling at the node (Camus et al., 2000; Schier and Shen, 2000; Vincent et al., 2003; Whitman, 2001). It is well established that signals emanating from the AME and anterior endoderm are required for patterning of the ventral neural tube and growth of the forebrain (Rubenstein, 2000). Lack of these signals may lead to cell death and defects in fore- and midbrain patterning. Furthermore, the AME has also been shown to regulate early expression of the Fgf8 at the MHB (Camus et al., 2000). In order to test whether the AME is preserved in Slb mutants, we analyzed the expression of Gsc, Shh and Foxa2, genes known to be expressed in the AME underlying the prospective forebrain (Belo et al., 1998; Chiang et al., 1996; Filosa et al., 1997). In Slb mutant embryos, Foxa2 expression is drastically reduced (n=4) and expression of both Gsc (n=5) and Shh (n=6) is absent (Fig. 8E–J).
Cells of the AME and the most anterior part of the ventral neural tube are derived from the posterior epiblast of the early primitive streak embryo (Tam and Behringer, 1997; Yamanaka et al., 2007). Thus observed defects in nodal expression and AME formation could conceivably be secondary to the defect in primitive streak formation. However, this is unlikely because the expression domain of Brachyury (T) in the Slb mutant embryos at E8.0 is similar to that in their wild type counterparts (data not shown). This indicates that the primitive streak is formed and elongates normally in the mutant. In sum the defect in AME formation is the earliest defect in brain development that we were able to detect in the Slb mutant.
DISCUSSION
Our study has shown that a targeted ablation of the Ift172 gene causes profound defects in brain development of the resulting Slb mutant embryo. The gene encodes a structural component of cilia. Thus, it is no surprise that the structure of these external cell organelles is severely affected in all brain regions examined, although the intracellular or basal portions of the cilia appear morphologically intact.
We noted an early onset and widespread distribution of Ift172 expression in the wild type post-implantation embryo. Impairment of embryonic development caused by targeted ablation of Ift172 function is early, severe, and ultimately lethal, affecting multiple vital organ systems. Forebrain truncations, holoprosencephaly and open cranial neural tubes are indications of severe rostral patterning defects. Reduction of Fgf8 in the anterior neural ridge and failure to maintain its expression in the developing forebrain leads to increased cell death and severe anterior truncation. In this context, the profound reduction in the expression of both Shh and its target Gli1 in the developing forebrain and midbrain is most notable, as also observed in the neural tube of the wimple mutant (Huangfu and Anderson, 2005; Huangfu et al., 2003). A high level of apoptosis in the developing Slb mutants as early as E10.0 precludes a more detailed analysis of the extent of defects in Shh expression, particularly in ventral aspects. However, given that impairment of Shh is drastic and widespread, the specification of the ventral brain is likely also affected.
Our analysis of anterior-posterior patterning in the developing mid- and hindbrain of the mutant reveals novel functions of Ift172 in the maintenance of the isthmic organizer, a signaling center formed between mid- and hindbrain that controls growth and patterning of this brain region. Previous studies have shown that Fgf8, Wnt1, and Pax2 form a regulatory loop essential for the formation and maintenance of the vertebrate isthmic organizer (Liu and Joyner, 2001), with Fgf8 assuming a key role in this process (Chi et al., 2003; Liu et al., 1999; Martinez et al., 1999). In the Slb mutant, Fgf8 is reduced dramatically at E8.3 at the MHB boundary. Consistent with this, expression of Gbx2, a direct target of Fgf8 (Liu et al., 1999) is substantially reduced in the first rhombomere. Wnt1 and Pax2 expression precedes that of Fgf8, and both factors have been shown to initiate Fgf8 at the MHB boundary, which in turn maintains their expression (McMahon and Bradley, 1990; McMahon et al., 1992; Ye et al., 2001; Schwarz et al., 1997). We find that Wnt1 and Pax2 expression in the Slb mutant at E8.5 is comparable to that of wild type embryos whereas, at E9.5 Wnt1 is reduced and the anterior expression domain of Pax2 is lost in the mutants. This loss of Wnt1 and Pax2 is most likely a consequence, rather than a cause, of the decreased Fgf8 levels. Fgf8 is down-regulated not only in Slb but also in Shh and SmoEn-1cre mutant embryos (Blaess et al., 2006; Zhang et al., 2001). However, it remains to be elucidated whether the reduction of Fgf8 expression at the MHB boundary that we observed in our Slb mutant as early as E8.5 reflects a defect in a feedback loop between Shh and Fgf8. Analysis of the Shh mutant cannot answer this question because early apoptosis precludes an analysis at this and subsequent stages of development. SmoEn-1cre mutants failed to maintain Fgf8 at MHB boundary at E11.0, but its level remained normal at E9.0 (Blaess et al., 2006) suggesting that regulation of early Fgf8 expression is most likely Hh/Smo independent.
The striking phenotypes of the developing mutant brain drew our attention to possible shortcomings in the function of signaling centers that organize brain morphogenesis and patterning prior to organogenesis in the gastrulating embryo. Our screen for signaling defects in the Slb mutant has revealed a dramatic reduction of nodal expression in the early streak stage embryo. As a member of the transforming growth factor beta (TGFβ) superfamily, nodal plays an essential role in early patterning events in gastrulating embryos (Shen, 2007). In the wild type embryo, this gene is initially expressed throughout the epiblast. In early streak embryos the actions of its antagonists Lefty1 and Cerberus-1 result in the restriction of nodal to the posterior epiblast (Yamamoto et al., 2004). In midstreak embryos, nodal mRNA is detected in the node as this embryonic organizer is formed at the anterior end of the primitive streak (Conlon et al., 1994; Zhou et al., 1993). Loss-of-function studies have revealed key roles of nodal in the establishment of the anterior-posterior (AP) axis, the induction of the primitive streak and the mesodermal and endodermal germ layers during gastrulation (Brennan et al., 2001; Norris et al., 2002).
Given the wealth of data showing an intimate connection between ciliogenesis and cell signaling events, there is good reason to believe that the defect in nodal expression observed in Slb mutants is closely related to cilia dysfunction in node precursor cells, thus affecting organizer function. This defect clearly affects the formation of a functional AME as reflected by the drastic reduction of the AME markers Shh, Gsc and Foxa2. Defects in AME formation have previously been noted in nodal mutants (Lowe et al., 2001; Vincent et al., 2003) further supporting the notion that the defect in nodal expression is a key element in the inability of the Slb mutant to establish functioning signaling centers required for brain patterning. Importantly, a severe reduction in nodal expression was registered in the epiblast as early as E7.0. Epiblast cells are not known to be ciliated. If this holds true, the Slb phenotype suggests a cilium-independent role of Ift172 in these cells. It is worthwhile to note, in this context, that immunostaining for IFT172 in Chlamydomonas has localized the protein not only to flagellae but also to cytoplasmic regions (Pedersen LB. et al., 2005).
Abnormal cilia morphogenesis, node dysfunction and failure to form normal AME ultimately lead to a defect in global brain patterning in Slb mutants. Signals from the AME are critical for growth and patterning of the forebrain. Furthermore, the mesendoderm of headfold stage embryos has also been shown to regulate early markers of the MHB boundary (Ang and Rossant, 1993; Camus et al., 2000). The resulting phenotypes, including truncation of anterior brain and holoprosencephaly, have also been observed in the nodal mutants cited above. Interestingly, mutations of genes in the nodal signaling pathway, including Cripto and TGIF, have been associated with holoprosencephaly in humans as well (de la Cruz et al., 2002; Gripp et al., 2000).
In conclusion, we have demonstrated that Ift172 plays a key role in cilia morphogenesis. We propose that Ift172 function in the node and/or epiblast is required for normal forebrain and mid-hindbrain patterning. Extensive efforts are underway to understand the mechanism that integrates cilia structure and function with the transduction of signals required for embryo development.
Supplementary Material
Acknowledgments
We thank Drs. P. Tam, M. Shen and A. Varela-Echavaria for helpful discussion and suggestions; A. Grinberg for assistance with embryonic stem cell cultures and blastocyst microinjection, R. Wu for help with genotyping, and Dr. C. Ott for helpful suggestions and technical assistance. This work was supported by funds from the intramural research program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
References
- Ang SL, Rossant J. Anterior mesendoderm induces mouse Engrailed genes in explant cultures. Development. 1993;118:139–149. doi: 10.1242/dev.118.1.139. [DOI] [PubMed] [Google Scholar]
- Belo JA, Leyns L, Yamada G, De Robertis EM. The prechordal midline of the chondrocranium is defective in Goosecoid-1 mouse mutants. Mech Dev. 1998;72:15–25. doi: 10.1016/s0925-4773(97)00204-9. [DOI] [PubMed] [Google Scholar]
- Bisgrove BW, Yost HJ. The roles of cilia in developmental disorders and disease. Development. 2006;133:4131–4143. doi: 10.1242/dev.02595. [DOI] [PubMed] [Google Scholar]
- Blaess S, Corrales JD, Joyner AL. Sonic hedgehog regulates Gli activator and repressor functions with spatial and temporal precision in the mid/hindbrain region. Development. 2006;133:1799–1809. doi: 10.1242/dev.02339. [DOI] [PubMed] [Google Scholar]
- Brennan J, Lu CC, Norris DP, Rodriguez TA, Beddington RS, Robertson EJ. Nodal signalling in the epiblast patterns the early mouse embryo. Nature. 2001;411:965–969. doi: 10.1038/35082103. [DOI] [PubMed] [Google Scholar]
- Camus A, Davidson BP, Billiards S, Khoo P, Rivera-Perez JA, Wakamiya M, Behringer RR, Tam PP. The morphogenetic role of midline mesendoderm and ectoderm in the development of the forebrain and the midbrain of the mouse embryo. Development. 2000;127:1799–1813. doi: 10.1242/dev.127.9.1799. [DOI] [PubMed] [Google Scholar]
- Chi CL, Martinez S, Wurst W, Martin GR. The isthmic organizer signal FGF8 is required for cell survival in the prospective midbrain and cerebellum. Development. 2003;130:2633–2644. doi: 10.1242/dev.00487. [DOI] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, Beachy PA. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- Conlon FL, Lyons KM, Takaesu N, Barth KS, Kispert A, Herrmann B, Robertson EJ. A primary requirement for nodal in the formation and maintenance of the primitive streak in the mouse. Development. 1994;120:1919–1928. doi: 10.1242/dev.120.7.1919. [DOI] [PubMed] [Google Scholar]
- Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- Cordes SP, Barsh GS. The mouse segmentation gene kr encodes a novel basic domain-leucine zipper transcription factor. Cell. 1994;79:1025–1034. doi: 10.1016/0092-8674(94)90033-7. [DOI] [PubMed] [Google Scholar]
- Crossley PH, Martin GR. The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development. 1995;121:439–451. doi: 10.1242/dev.121.2.439. [DOI] [PubMed] [Google Scholar]
- Davenne M, Maconochie MK, Neun R, Pattyn A, Chambon P, Krumlauf R, Rijli FM. Hoxa2 and Hoxb2 control dorsoventral patterns of neuronal development in the rostral hindbrain. Neuron. 1999;22:677–691. doi: 10.1016/s0896-6273(00)80728-x. [DOI] [PubMed] [Google Scholar]
- de la Cruz JM, Bamford RN, Burdine RD, Roessler E, Barkovich AJ, Donnai D, Schier AF, Muenke M. A loss-of-function mutation in the CFC domain of TDGF1 is associated with human forebrain defects. Hum Genet. 2002;110:422–428. doi: 10.1007/s00439-002-0709-3. [DOI] [PubMed] [Google Scholar]
- Filosa S, Rivera-Perez JA, Gomez AP, Gansmuller A, Sasaki H, Behringer RR, Ang SL. Goosecoid and HNF-3beta genetically interact to regulate neural tube patterning during mouse embryogenesis. Development. 1997;124:2843–2854. doi: 10.1242/dev.124.14.2843. [DOI] [PubMed] [Google Scholar]
- Gavalas A, Davenne M, Lumsden A, Chambon P, Rijli FM. Role of Hoxa-2 in axon pathfinding and rostral hindbrain patterning. Development. 1997;124:3693–3702. doi: 10.1242/dev.124.19.3693. [DOI] [PubMed] [Google Scholar]
- Gripp KW, Wotton D, Edwards MC, Roessler E, Ades L, Meinecke P, Richieri-Costa A, Zackai EH, Massague J, Muenke M, Elledge SJ. Mutations in TGIF cause holoprosencephaly and link NODAL signalling to human neural axis determination. Nat Genet. 2000;25:205–208. doi: 10.1038/76074. [DOI] [PubMed] [Google Scholar]
- Hagiwara H, Ohwada N, Aoki T, Takata K. Ciliogenesis and ciliary abnormalities. Med Electron Microsc. 2000;33:109–114. doi: 10.1007/s007950000009. [DOI] [PubMed] [Google Scholar]
- Haycraft CJ, Banizs B, Aydin-Son Y, Zhang Q, Michaud EJ, Yoder BK. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard PW, Maurer RA. Identification of a conserved protein that interacts with specific LIM homeodomain transcription factors. J Biol Chem. 2000;275:13336–13342. doi: 10.1074/jbc.275.18.13336. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Anderson KV. Cilia and Hedgehog responsiveness in the mouse. Proc Natl Acad Sci U S A. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Anderson KV. Signaling from Smo to Ci/Gli: conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development. 2006;133:3–14. doi: 10.1242/dev.02169. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- Jones C, Roper VC, Foucher I, Qian D, Banizs B, Petit C, Yoder BK, Chen P. Ciliary proteins link basal body polarization to planar cell polarity regulation. Nat Genet. 2008;40:69–77. doi: 10.1038/ng.2007.54. [DOI] [PubMed] [Google Scholar]
- Kulaga HM, Leitch CC, Eichers ER, Badano JL, Lesemann A, Hoskins BE, Lupski JR, Beales PL, Reed RR, Katsanis N. Loss of BBS proteins causes anosmia in humans and defects in olfactory cilia structure and function in the mouse. Nat Genet. 2004;36:994–998. doi: 10.1038/ng1418. [DOI] [PubMed] [Google Scholar]
- Liu A, Joyner AL. Early anterior/posterior patterning of the midbrain and cerebellum. Annu Rev Neurosci. 2001;24:869–896. doi: 10.1146/annurev.neuro.24.1.869. [DOI] [PubMed] [Google Scholar]
- Liu A, Losos K, Joyner AL. FGF8 can activate Gbx2 and transform regions of the rostral mouse brain into a hindbrain fate. Development. 1999;126:4827–4838. doi: 10.1242/dev.126.21.4827. [DOI] [PubMed] [Google Scholar]
- Lowe LA, Yamada S, Kuehn MR. Genetic dissection of nodal function in patterning the mouse embryo. Development. 2001;128:1831–1843. doi: 10.1242/dev.128.10.1831. [DOI] [PubMed] [Google Scholar]
- Manley NR, Capecchi MR. The role of Hoxa-3 in mouse thymus and thyroid development. Development. 1995;121:1989–2003. doi: 10.1242/dev.121.7.1989. [DOI] [PubMed] [Google Scholar]
- Martinez S, Crossley PH, Cobos I, Rubenstein JL, Martin GR. FGF8 induces formation of an ectopic isthmic organizer and isthmocerebellar development via a repressive effect on Otx2 expression. Development. 1999;126:1189–1200. doi: 10.1242/dev.126.6.1189. [DOI] [PubMed] [Google Scholar]
- May SR, Ashique AM, Karlen M, Wang B, Shen Y, Zarbalis K, Reiter J, Ericson J, Peterson AS. Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Dev Biol. 2005;287:378–389. doi: 10.1016/j.ydbio.2005.08.050. [DOI] [PubMed] [Google Scholar]
- McMahon AP, Bradley A. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell. 1990;62:1073–1085. doi: 10.1016/0092-8674(90)90385-r. [DOI] [PubMed] [Google Scholar]
- McMahon AP, Joyner AL, Bradley A, McMahon JA. The midbrain-hindbrain phenotype of Wnt-1-/Wnt-1- mice results from stepwise deletion of engrailed-expressing cells by 9.5 days postcoitum. Cell. 1992;69:581–595. doi: 10.1016/0092-8674(92)90222-x. [DOI] [PubMed] [Google Scholar]
- Norris DP, Brennan J, Bikoff EK, Robertson EJ. The Foxh1-dependent autoregulatory enhancer controls the level of Nodal signals in the mouse embryo. Development. 2002;129:3455–3468. doi: 10.1242/dev.129.14.3455. [DOI] [PubMed] [Google Scholar]
- Pan J, Wang Q, Snell WJ. Cilium-generated signaling and cilia-related disorders. Lab Invest. 2005;85:452–463. doi: 10.1038/labinvest.3700253. [DOI] [PubMed] [Google Scholar]
- Park TJ, Haigo SL, Wallingford JB. Ciliogenesis defects in embryos lacking inturned or fuzzy function are associated with failure of planar cell polarity and Hedgehog signaling. Nat Genet. 2006;38:303–311. doi: 10.1038/ng1753. [DOI] [PubMed] [Google Scholar]
- Pedersen LB, Miller MS, Geimer S, Leitch JM, Rosenbaum JL, Cole DG. Chlamydomonas IFT172 is encoded by FLA11, interacts with CrEB1, and regulates IFT at the flagellar tip. Curr Biol. 2005;15:262–266. doi: 10.1016/j.cub.2005.01.037. [DOI] [PubMed] [Google Scholar]
- Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Ross AJ, May-Simera H, Eichers ER, Kai M, Hill J, Jagger DJ, Leitch CC, Chapple JP, Munro PM, Fisher S, Tan PL, Phillips HM, Leroux MR, Henderson DJ, Murdoch JN, Copp AJ, Eliot MM, Lupski JR, Kemp DT, Dollfus H, Tada M, Katsanis N, Forge A, Beales PL. Disruption of Bardet-Biedl syndrome ciliary proteins perturbs planar cell polarity in vertebrates. Nat Genet. 2005;37:1135–1140. doi: 10.1038/ng1644. [DOI] [PubMed] [Google Scholar]
- Rubenstein JL. Intrinsic and extrinsic control of cortical development. Novartis Found Symp. 2000;228:67–75. doi: 10.1002/0470846631.ch6. discussion 75–82, 109–113. [DOI] [PubMed] [Google Scholar]
- Schier AF, Shen MM. Nodal signalling in vertebrate development. Nature. 2000;403:385–389. doi: 10.1038/35000126. [DOI] [PubMed] [Google Scholar]
- Schneider-Maunoury S, Topilko P, Seitandou T, Levi G, Cohen-Tannoudji M, Pournin S, Babinet C, Charnay P. Disruption of Krox-20 results in alteration of rhombomeres 3 and 5 in the developing hindbrain. Cell. 1993;75:1199–1214. doi: 10.1016/0092-8674(93)90329-o. [DOI] [PubMed] [Google Scholar]
- Schneider L, Clement CA, Teilmann SC, Pazour GJ, Hoffmann EK, Satir P, Christensen ST. PDGFRalphaalpha signaling is regulated through the primary cilium in fibroblasts. Curr Biol. 2005;15:1861–1866. doi: 10.1016/j.cub.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Scholey JM, Anderson KV. Intraflagellar transport and cilium-based signaling. Cell. 2006;125:439–442. doi: 10.1016/j.cell.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Schwarz M, Alvarez-Bolado G, Urbanek P, Busslinger M, Gruss P. Conserved biological function between Pax-2 and Pax-5 in midbrain and cerebellum development: evidence from targeted mutations. Proc Natl Acad Sci U S A. 1997;94:14518–14523. doi: 10.1073/pnas.94.26.14518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MM. Nodal signaling: developmental roles and regulation. Development. 2007;134:1023–1034. doi: 10.1242/dev.000166. [DOI] [PubMed] [Google Scholar]
- Sun Z, Amsterdam A, Pazour GJ, Cole DG, Miller MS, Hopkins N. A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development. 2004;131:4085–4093. doi: 10.1242/dev.01240. [DOI] [PubMed] [Google Scholar]
- Tam PP, Behringer RR. Mouse gastrulation: the formation of a mammalian body plan. Mech Dev. 1997;68:3–25. doi: 10.1016/s0925-4773(97)00123-8. [DOI] [PubMed] [Google Scholar]
- Tsujikawa M, Malicki J. Intraflagellar transport genes are essential for differentiation and survival of vertebrate sensory neurons. Neuron. 2004;42:703–716. doi: 10.1016/s0896-6273(04)00268-5. [DOI] [PubMed] [Google Scholar]
- Vincent SD, Dunn NR, Hayashi S, Norris DP, Robertson EJ. Cell fate decisions within the mouse organizer are governed by graded Nodal signals. Genes Dev. 2003;17:1646–1662. doi: 10.1101/gad.1100503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman M. Nodal signaling in early vertebrate embryos: themes and variations. Dev Cell. 2001;1:605–617. doi: 10.1016/s1534-5807(01)00076-4. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG, Bhatt S, Chavrier P, Bravo R, Charnay P. Segment-specific expression of a zinc-finger gene in the developing nervous system of the mouse. Nature. 1989;337:461–464. doi: 10.1038/337461a0. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Saijoh Y, Perea-Gomez A, Shawlot W, Behringer RR, Ang SL, Hamada H, Meno C. Nodal antagonists regulate formation of the anteroposterior axis of the mouse embryo. Nature. 2004;428:387–392. doi: 10.1038/nature02418. [DOI] [PubMed] [Google Scholar]
- Yamanaka Y, Tamplin OJ, Beckers A, Gossler A, Rossant J. Live imaging and genetic analysis of mouse notochord formation reveals regional morphogenetic mechanisms. Dev Cell. 2007;13:884–896. doi: 10.1016/j.devcel.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Ye W, Bouchard M, Stone D, Liu X, Vella F, Lee J, Nakamura H, Ang SL, Busslinger M, Rosenthal A. Distinct regulators control the expression of the mid-hindbrain organizer signal FGF8. Nat Neurosci. 2001;4:1175–1181. doi: 10.1038/nn761. [DOI] [PubMed] [Google Scholar]
- Zhang XM, Ramalho-Santos M, McMahon AP. Smoothened mutants reveal redundant roles for Shh and Ihh signaling including regulation of L/R symmetry by the mouse node. Cell. 2001;106:781–792. [PubMed] [Google Scholar]
- Zhou X, Sasaki H, Lowe L, Hogan BL, Kuehn MR. Nodal is a novel TGF-beta-like gene expressed in the mouse node during gastrulation. Nature. 1993;361:543–547. doi: 10.1038/361543a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.