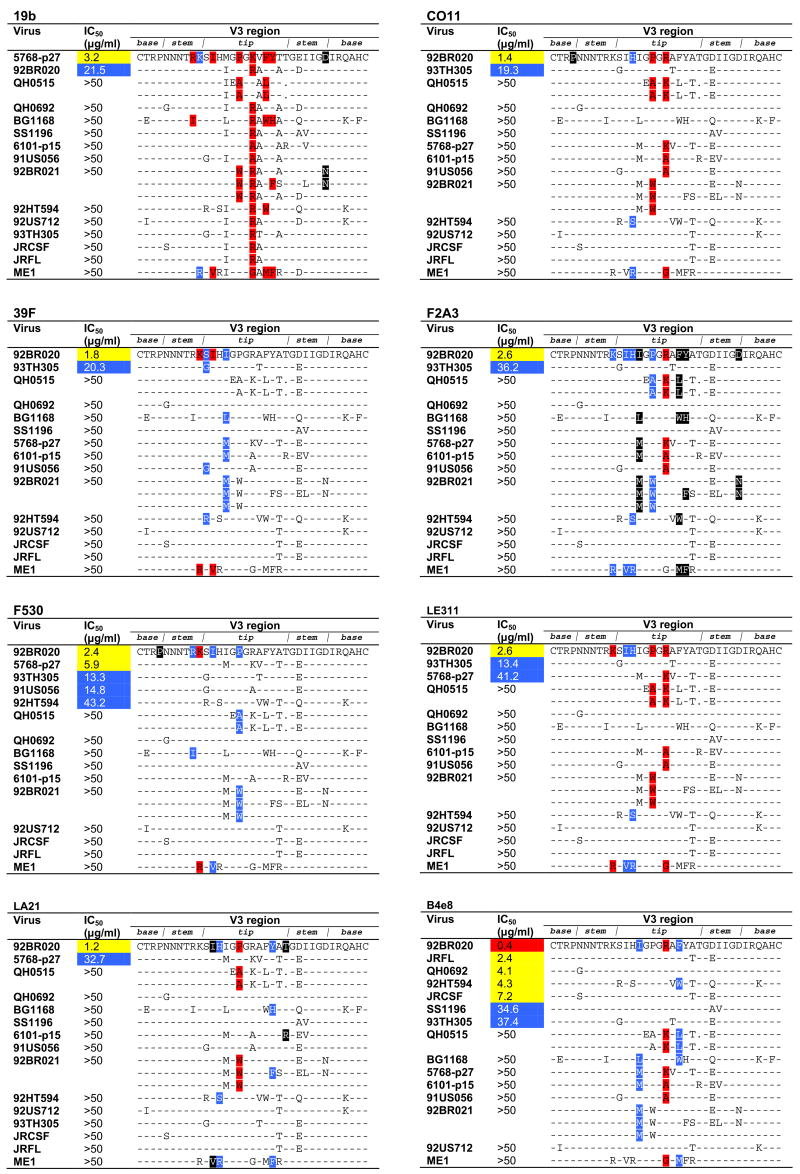
Fig. 1.
Epitope specificity of V3 mAbs related to neutralizing activity. The V3 sequences of the 15 subtype B viruses investigated in this study are shown. For viruses QH0515 and 92BR021, multiple sequences are listed to represent the different V3 sequences that have been reported from quasispecies pools of these 2 viruses; for the other viruses for which quasispecies pools were used only a single sequence (shown) has been reported. To depict which residues are most permissible for each mAb, viruses are ranked according to their neutralization sensitivity; viruses not neutralized are ranked in the order as they appear in Table 1. The locations of those residues that substantially diminish or enhance mAb binding are colored as in Table 2; dashes indicate sequence homology with the virus that was neutralized most potently and, to maintain clarity, are not colored. Sequence gaps relative to the V3 sequence of the most potently neutralized virus are denoted by a dot. Sequence alignment was performed using ClustalW2 (http://www.ebi.ac.uk/clustalw/) (Larkin et al., 2007) and formatted for publication using the sequence alignment publishing tool SeqPublish (http://www.hiv.lanl.gov/content/hiv-db/SeqPublish/seqpublish.html).