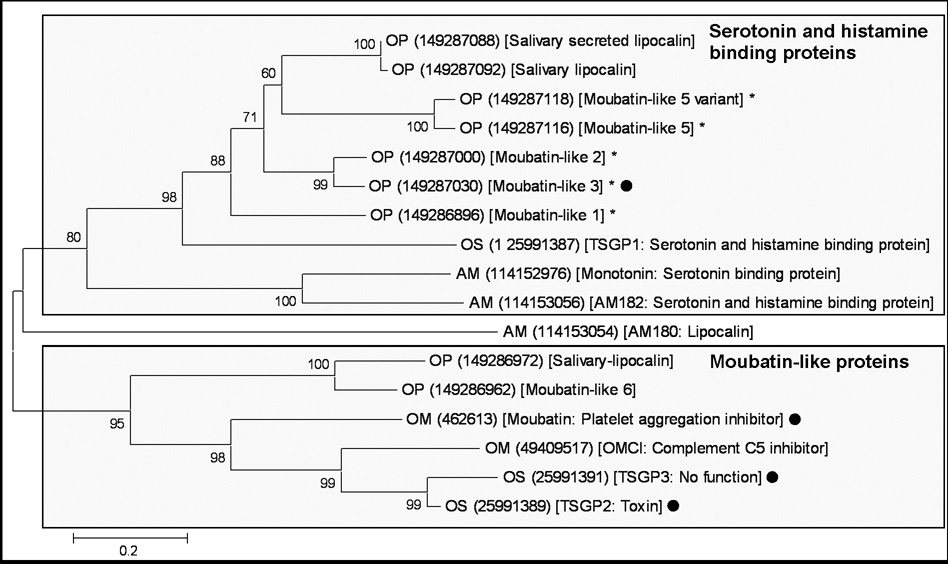
Fig. 1.
Phylogenetic analysis of moubatin-related lipocalins. Lipocalins were retrieved from the non-redundant protein database using the sequence for moubatin. All hits with E-values below one were extracted and used to construct a neighbor-joining tree. Support values (10 000 bootstraps) for nodes is indicated. The two major supported clades are boxed. Sequences are identified by a species designation (AM: Argas monolakensis; OM: Ornithodoros moubata; OP: O. parkeri, OS: O. savignyi) and a Genbank accession number within round brackets. The Genbank description line and functional information extracted from the literature is included within square brackets. Sequences marked with dots were characterized in the present study. Those marked with asterisks indicate proteins whose functional and evolutionary relationship with moubatin is uncertain.