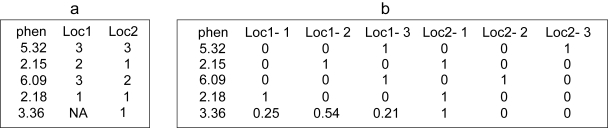
Figure 1. Illustration of data formatting.
Part a provides an example of a data set in which the genotypes of individals are fully known (or, alternatively, totally unknown and considered as missing data); 1 and 3 stand for the homozygotes (e.g. ‘AA’ and ‘aa’) and 2 for the hererozygote. Part b illustrates a second kind of data set in which the genotypes are defined by their probabilites. In this example, part b is the exact equivalent of part a (and then, the frequency of the ‘known’ genotypes is always 1), but in practice, especially when the data result from a Haley-Knott regression, the probabilities, computed from the genotypes at flanking markers, may be intermediate. Missing values (‘NA’) are allowed in type a data sets, and are replaced by genotypic probabilities equal to genotypic frequencies in the rest of the population (here, close to 0.25, 0.5, and 0.25 since the population is an F2). The Z matrix used for the regression (equation 5) is computed from a ‘type b’ data set, meaning that if ‘type a’ data is provided, it is turned into ‘type b’ before the genetic regression.