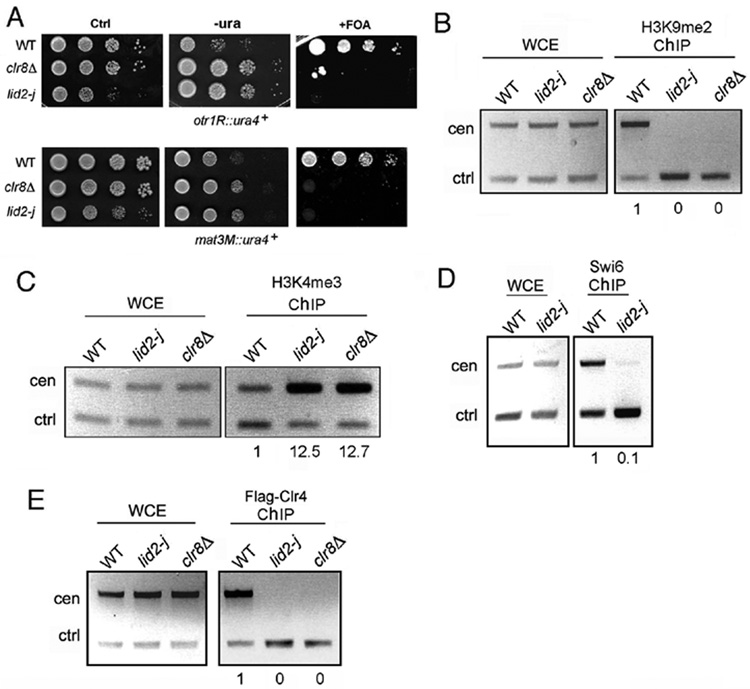
Figure 3. Lid2 is required for heterochromatin silencing.
A) Heterochromatin silencing is lost in lid2-j as shown by a silencing assay. lid2-j and clr8Δ containing the ura4+ marker at a otr (upper panel) or mating-type region (lower panel) were plated using a serial dilution on minimal medium without uracil or with the counter-selective FOA media. Ctrl, a control plate containing uracil.
B) H3K9 methylation is lost at centromeres in lid2-j. A ChIP assay was carried out with the indicated strains using the antibody against H3K9me2. The centromere fragment from otr region and control (alp5+) were amplified from immunoprecipitated DNA by multiplex PCR. WCE, whole-cell extracts before immunoprecipitation; ChIP, immunoprecipitated DNA. The ratios of heterochromatin signals versus control signals between ChIP and WCE fractions were used to calculate the relative enrichment of precipitated DNA, shown below each lane. The value for WT was set to 1.0.
C) H3K4 methylation is increased in heterochromatin in lid2-j. ChIP assay was performed as in B) using the antibody against H3K4me3.
D) Swi6 binding is abolished at the centromere in lid2-j as shown by ChIP assay using anti-Swi6 antibody.
E) Lid2 is required for recruitment of Clr4 to the centromere. A ChIP assay was carried out with the indicated strains carrying Flag-Clr4 using the antibody against Flag.