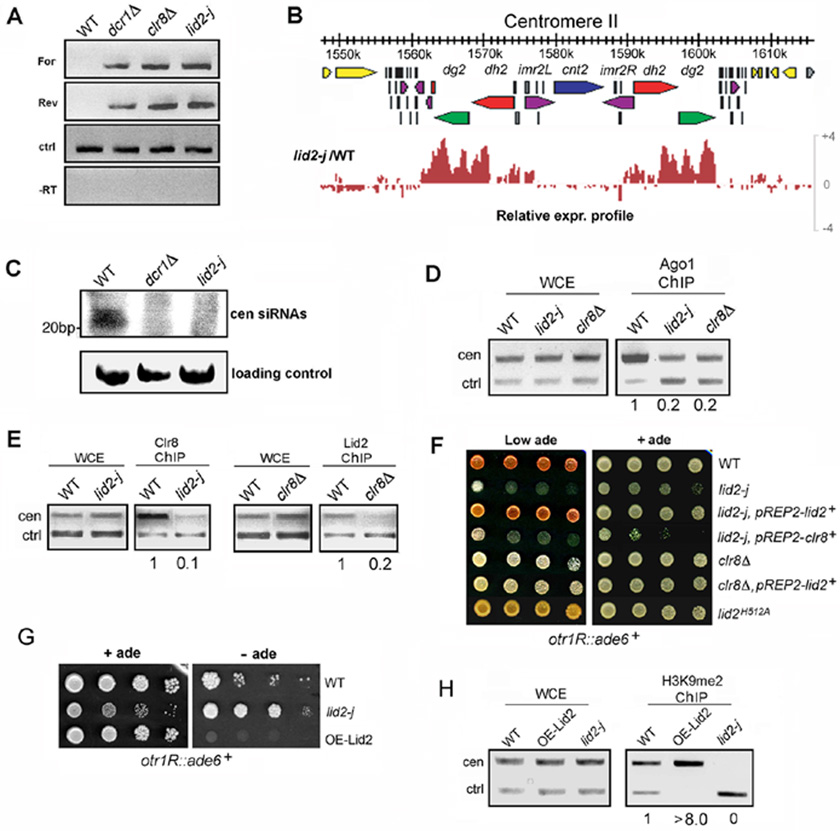
Figure 5. Lid2 acts in the RNAi pathway and regulates Clr4 methylation in a dose-dependent manner by functioning cooperatively with Clr8.
A) Centromere transcripts accumulated in lid2-j as demonstrated using strand-specific RT-PCR. alp5+ serves as a control (ctrl). For, forward centromere transcript; Rev, reverse centromere transcript; -RT, no reverse transcriptase.
B) The genomic expression distribution at centromere II in lid2-j is shown using Genome Browser analysis. Genes (yellow and gray), dg repeats (green), dh repeats (red), imr repeats (purple), and tRNA genes (black) are indicated by different colors. Note that transcripts from heterochromatin region are significantly increased in lid2-j.
C) A small RNA northern blot showed that centromeric siRNA was reduced in lid2-j. Small RNAs spanning 22 to 26 nucleotides corresponding to the centromeric dh repeat were readily detected in the WT. These small RNAs were lost in the lid2 and dcr1 mutants.
D) The association of Ago1 with the centromere is reduced in lid2-j. A ChIP assay was conducted in WT and lid2-j using antibody against Ago1. The centromere fragment from otr region and control (alp5+) were amplified.
E) Clr8 and Lid2 mutually affect each other’s recruitment to heterochromatin. ChIP was carried out for lid2-j and WT containing Myc-Clr8 using antibody against the Myc tag. ChIP also was performed for clr8Δ and WT containing Lid2-TAP using IgG sepharose. The centromere fragment and control (alp5+) were amplified.
F) Expressing Lid2 in clr8Δ can not rescue the mutant’s silencing defect, and neither can expressing Clr8 in lid2-j; the silencing assays also show that the point mutant, lid2H512A, can cause significant loss of silencing. The strains indicated containing the ade6+ marker in the otr region were plated by a serial dilution on a YES medium without supplementing adenine. Repression of ade6+ expression results in pink color.
G) Overexpression of Lid2 significantly enhanced heterochromatic silencing. The strains containing ade6+ in the otr region were plated on a minimal medium lacking adenine.
H) H3K9 methylation in the otr region was increased in the strain overexpressing Lid2. ChIP assay was performed as in Figure 3B.