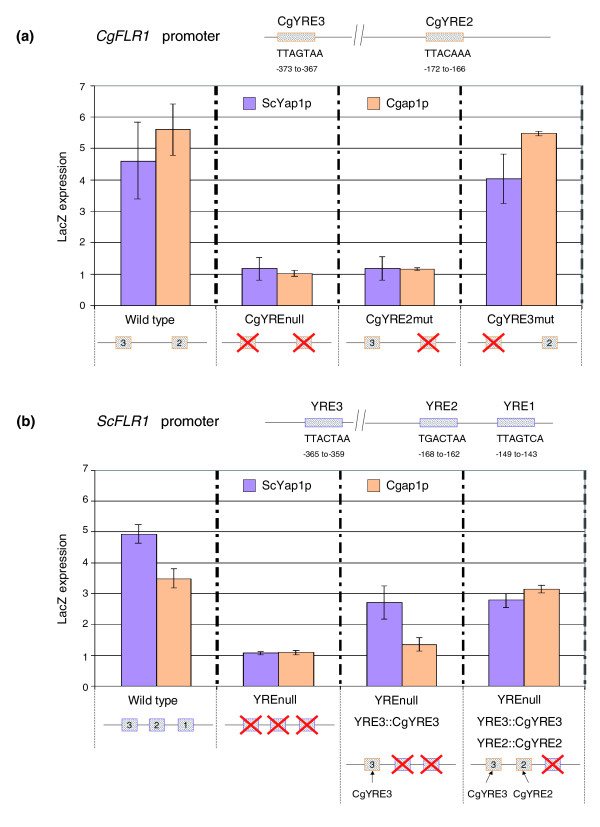
Figure 4.
Functional comparative analyses of ScYap1p and Cgap1p activities in vivo. In vivo assays of ScYap1p and Cgap1p properties were conducted, using S. cerevisiae strains expressing either ScYap1p (purple histograms) or Cgap1p (orange histograms). LacZ was used as a reporter gene and was placed under the control of wild-type or mutated versions of (a) the CgFLR1 or (b) ScFLR1 promoter regions (see Materials and methods). Descriptions of the mutations performed in YREs are shown in Additional data file 12. LacZ expression was measured by real-time quantitative RT-PCR, before and after benomyl treatment (20 μg/ml) for 40 minutes. (a) Only the inactivation of CgYRE2 (TTACAAA) dramatically decreased the benomyl response of CgFLR1. In the context of a C. glabrata promoter (in this case, CgFLR1) TTACAAA acts as the major BRE. (b) The LacZ reporter gene was placed under the control of the ScFLR1 promoter, in which all three YREs were inactivated and replaced with CgYRE3 and CgYRE2 sequences. To summarize, ScYap1p appeared to be as efficient at the ScFLR1 and at the CgFLR1 wild-type promoters, whereas Cgap1p was more efficient at the CgFLR1 promoter (a, b). Moreover, only the introduction of CgYRE2 was able to restore the full activity of Cgap1p at the ScFLR1 mutated promoter, whereas the sole introduction of CgYRE3 sequence restored half of the ScYap1p activity, and the addition of the CgYRE2 sequence did not increase this activity. In the heterologous context of the ScFLR1 promoter, CgYRE2 is still the main BRE for Cgap1p, but not for ScYap1p, which prefers CgYRE3.