Abstract
Expression microarrays identified a novel transcript, designated as Ugene, whose expression is absent in normal colon and colon adenomas, but that is commonly induced in malignant colon cancers. These findings were validated by real-time PCR and Northern blot analysis in an independent panel of colon cancer cases. In addition, Ugene expression was found to be elevated in many other common cancer types, including, breast, lung, uterus, and ovary. Immunofluorescence of V5-tagged Ugene revealed it to have a nuclear localization. In a pull-down assay, uracil DNA-glycosylase 2 (UNG2), an important enzyme in the base excision repair pathway, was identified as a partner protein that binds to Ugene. Co-immunoprecipitation and Western blot analysis confirmed the binding between the endogenous Ugene and UNG2 proteins. Using deletion constructs, we find that Ugene binds to the first 25 amino acids of the UNG2 NH2-terminus. We suggest Ugene induction in cancer may contribute to the cancer phenotype by interacting with the base excision repair pathway.
Keywords: Ugene, colon cancer, UNG2, base excision repair
Introduction
Cancers of the colon and rectum (CRC) are the second leading cause of cancer deaths among adult Americans. Colon cancer develops as a result of the progressive accumulation of genetic and epigenetic alterations in key onco- and tumor suppressor- genes that lead to the transformation of normal colonic epithelium to adenocarcinoma (1). In addition to primary genetic alterations, the cancer phenotype is also importantly modulated by down stream alterations in the levels of expression of different effector genes (2–4). Expression microarrays enable comprehensive profiling of the cancer transcriptome and identification of cancer specific changes in gene expression (5, 6).
In this study, using a global gene expression profiling array, we identified a previously uncharacterized gene, referred to as Ugene, which is overexpressed in malignant colon cancers. We further demonstrate that Ugene is frequently elevated in most malignant tumor types. In addition, we provide experimental evidence showing that Ugene protein is localized within the nucleus and forms a complex with uracil DNA glycosylase 2 (UNG2), a base excision repair (BER) enzyme.
Materials and Methods
Cell lines and tissues
VACO cell lines were established and maintained as previously described (7). DLD1 and SW480 cell lines were obtained from American Type Culture Collection (ATCC, Manassas, VA). Normal colons, primary colon cancers, and liver metastasis tissues were obtained from the archives of University Hospitals of Cleveland (Cleveland, OH) under an IRB-approved protocol. Total RNA and genomic DNA were prepared as described (8).
DNA expression microarray analysis
As described previously (9), we designed custom expression monitoring microarrays using Affymetrix GeneChip technology (10). Preparation of samples, hybridization to GeneChip expression microarrays, and data analysis were all performed as described previously (9).
Rapid amplification of cDNA ends (RACE)-PCR
5′ and 3′ RACE-ready cDNA was generated from 2μg of total RNA (V241 cell line) using the 5′/3′ RACE kit (Roche, Indianapolis, IN). The gene-specific primers used for 5′ RACE were as follows: SP1, 5′-GCG GGA CCT AGA GCT TTT CT-3′; SP2, 5′-GAG GCA GGT GGA GTT TGA AG-3′; and SP3, 5′-ATC CCT TCC CCA GCA TTA AG-3′. The gene-specific primer for 3′ RACE was: 5′-ACC TCA TCC TTC CTG CGA CG-3′. Full-length Ugene was PCR amplified from RACE-ready cDNA using the forward 5′-CCG ACT GAG CCT CTA AAG CGA C-3′ and reverse 5′-TCC TGA TTC ACA AAC TCT TGC TCC-3′ primers.
Northern analysis
Northern analysis was performed as previously described (11) using the entire Ugene coding region as the probe.
Southern analysis
Total genomic DNA from cell lines and normal tissues were digested with PstI, separated by electrophoresis on a 0.8% agarose gel and transferred onto a Zeta-Probe blotting membrane. 32P labeled DNA probes were prepared by random primer extension of a fragment containing the Ugene coding sequence. Equal loading of DNA was confirmed by rehybridizing blots with a probe designated to the TGFβ-RII gene, which is relatively copy number invariant in CRC.
Human cancer dot blots
Radioactively labeled cDNA probes were synthesized from human Ugene or ubiquitin control cDNA using random primer labeling followed by probe purification on CHROMA SPIN+STE-100 columns (BD Biosciences). Hybridization of the Cancer Profiling Array with human Ugene probes and washings of the array were done according to the manufacturer’s recommendations (BD Biosciences). The hybridized Cancer Profiling Arrays were then exposed to the phosphorimaging screens and scanned with a Storm 840 PhosphorImager. We then stripped this same membrane and hybridized it with human ubiquitin cDNA probe to show equal sample loading.
Ugene real-time PCR
Primers and a fluorogenic hybridization probe were designed using Primer3 software (12). Ugene was amplified using 400 nM of forward primer 5′-CTG TCT TCT TTC CTG CAA CAA C-3′ and reverse primer 5′-TAG GAC GTT TAC ACC TGT GGA G-3′, and detected using fluorogenic hybridization probe 5′-/56-FAM/ATA AAC TGC CTG GCT GTG AAA CAT CCA G/3BHQ_2/-3′. Beta-2-microglobulin (B2M) was amplified using 0.2× of the human B2M TaqMan primer/probe kit (Perkin-Elmer Biosciences, Foster City, CA). Each PCR was carried out in triplicate in a 25 μL volume using TaqMan Assay Mastermix (Applied Biosystems, Foster City, CA) for 8 min at 95°C, followed by 50 cycles of 95°C for 15 s, 57°C for 30 s, and 72°C for 30 s. The level of Ugene expression was determined as the ratio of Ugene: B2M = 2 (CT B2M-CT Ugene).
Construction of expression/deletion vectors
The coding sequence of Ugene (Ugene-p/Ugene-q, XM_001133365) and UNG2 (NM_080911) was PCR amplified and cloned into the eukaryotic expression vector pcDNA3.1/V5/His-TOPO (Invitrogen, Carlsbad, CA) to generate COOH-terminal V5-tagged Ugene/UNG2 expression vectors. The primer sequences for constructing the vectors are as follows: for Ugene, forward 5′-ACC TCA TCC TTC CTG CGA CG-3′ and reverse 5′-TCT AAT ACA CTC CTC TGC TGA GAT-3′; for UNG2, forward 5′-ATG GGC GTC TTC TGC CTT G-3′ and reverse 5′-CAG CTC CTT CCA GTC AAT G-3′. FLAG tagged constructs were similarly made by adding the complimentary FLAG tag sequence with a stop codon (5′-TTA CTT GTC ATC GTC GTC CTT GTA GTC-3′) at the 5′ end of the reverse primer. UNG2 deletion constructs were generated by blunt-ligation of the PCR products, amplified using V5-tagged UNG2 expression vector as a template, with the primer sets as listed in supplemental table 1. The decoy fusion protein 1-25-UNG2-GFP expression vector was constructed by ligating the following 3 fragments: a) green fluorescence protein (GFP) DNA [that was PCR amplified from pEGFP-N1 template (Clontech, Mountain View, CA) using forward 5′-TTG AAT TCA TGG TGA GCA AGG GCG AGG AG-3′ and reverse 5′-TTC TCG AGC CCT TGT ACA GCT CGT CCA TGC-3′, and digested with EcoRI and XhoI]; b) nucleotides corresponding to 1-25 amino acids of UNG2 (that were amplified using forward 5′-TTG GAT CCC TCC TCA GCT CCA GGA TGA T-3′ and reverse 5′-TTG AAT TCC TCG GGG CTG GGG GCG TGT-3′, then digested with BamH1 and EcoRI); c) pcDNA4/myc-His(B) (Invitrogen) fragment (that was obtained by digesting the vector with BamHI and XhoI). In this construct, 1-25-UNG2-GFP is driven by a CMV promoter with two copies of the TetO2 operator sequence, which can be suppressed by the Tet repressor.
Construction of DLD1 clones conditionally expressing 1-25-UNG2-GFP
DLD1 cells were seeded at 1.0×106 cells/100mm dish and transfected the next day with 1.6 μg of pcDNA6/TR and 0.4μg pcDNA4/myc-His/1-25-UNG2-GFP plasmid using 12 μL of Fugene 6 (Roche Applied Sciences) as per the manufacturer’s protocols, then selected by blasticidine (10μg/ml) and zeocin (200μg/ml) for 2 weeks to derive clones conditionally expressing 1-25-UNG2-GFP fusion protein under doxycycline (0.5μg/mL) regulation.
Immunofluorescence
SW480 cells were seeded at 1.0×106 cells/100mm dish and transfected the next day with 2 μg of V5-tagged Ugene expression vector using 12 μL of Fugene 6 (Roche Applied Sciences, Indianapolis, IN) as per the manufacturer’s protocols. Immunofluorescence was performed 48 hours after transfection using V5 antibody (Invitrogen) at 1:200, followed by Alexa Fluor 488 goat anti mouse IgG antibody (Invitrogen) at 1:400.
Immunoprecipitation and Western blot analysis
HEK 293T cells were seeded at 4.0×106 cells/T75 flask and transfected the next day with total 6 μg of plasmids using 36 μL of LipofectAMINE (Invitrogen). Cell lysates were prepared 48 hours after tranfection using lysis buffer (50 mM Tris-HCl pH7.5, 1 mM EDTA pH8.0, 150 mM NaCl, 1% Triton X-100) supplemented with protease inhibitor mixture (Roche Applied Sciences). FLAG immunoprecipitation was performed with anti-FLAG M2 affinity gel as described in the manufacturer’s protocol. After elution by either FLAG-peptide (Sigma, St. Louis, MO) or 3xFLAG peptide (Sigma), eluates were used for biochemical activity assay or for Western blot. Western blots were performed using anti-FLAG M2 (Sigma, 1:1000), anit-V5 (1:1000, Invitrogen), anti-UNG (1:500, Abcam, Cambridge, MA), anti-GFP (1:5000, Invitrogen), or anti-beta actin (1:100,000, Sigma) antibodies, followed by horseradish peroxide conjugated donkey anti-mouse secondary antibody (1:1500, Jackson ImmunoResearch Laboratories, Grove City, PA), and visualized by using an Enhanced Chemiluminescence Plus detection kit (Amersham Biosciences).
Somatic cell knockout
Somatic cell knockout was performed as described (13). Knockout of both UNG1 and UNG2 transcription units was accomplished by disrupting UNG locus exon 2 using left and right targeting arms amplified with primers: left arm 5′-GGC TCG AGA GGC ACA AAG CGA ATG AAA G-3′, 5′-CCG AAT TCA GTC AGT CAC TCT GGA TCC GGT CCA ACT-3′, right arm 5′-GGA CTA GTG GAG AGA GCT GGA AGA AGC A-3′, 5′-AAC CGC GGT TTG AAC TTC ACC ACC ACC A-3′. UNG1 specific knockout was accomplished by eliminating the UNG1 specific exon 1 using targeting arms amplified with primers: left arm 5′-GGC TCG AGA AGA GCC TGT CCA AAG AGC A-3′/5′-CCG AAT TCC GGG AAT TGG GAA TTA GGT T-3′, right arm 5′-GGA CTA GTC TCT TGA GCC GCC TCT GC-3′/5′-AAC CGC GGT TTG AAC TTC ACC ACC ACC A-3′. Cells expressing only UNG2 were generated by UNG1 specific knockout of one allele combined with the UNG locus knockout of the second allele.
Endogenous Ugene epitope tagging
3xFLAG tagging of the endogenous Ugene-p gene was performed by somatic cell knock-in vectors as described in Zhang et al, 2008 (14). The primers used were: left arm 5′-GGC TCG AGC AAC CTG GCC CTA AAG TTC A-3′, 5′-CCG ATA TCT CTA ATA CAC TCC TCT GCT GAG-3′, right arm 5′-GGA CTA GTA TGG AAT TAT GAT ATA TAT GAT ATA C-3′, 5′-AAC CGC GGC AAA ACC ACA ACT CAG TCT GCT-3′.
siRNA-mediated Ugene silencing
The Ugene-specific and control siRNAs were synthesized by Dharmacon. For siRNA transfection experiments, DLD1 cells were seeded on 100 mm culture dishes and transfected with 30 μL of 20 μM siRNA stock using 30 μL LipofectAMINE 2000 (Invitrogen). Cell lysates were collected 48 hours after transfection and knockdown was validated by Western blot. The sequences of Ugene siRNA-1017 were as follows: sense 5′-GGA AGA UGC UAU UUC ACC AUU-3′, antisense 5′-pUGG UGA AAU AGC AUC UUC CUU-3′.
In vitro UNG biochemical activity assay
The uracil-containing oligonucleotide (5′-CCT GCC CTG UGC AGC TGT GGG-3′) (R&D, Minneapolis, MN) was annealed to an equimolar amount of its complementary strand (5′-CCC ACA GCT GCA CAG GGC AGG-3′ or 5′-CCC ACA GCT GCG CAG GGC AGG-3′, for U:A and U:G pairs respectively), mixed and heated to 95°C in annealing buffer (20 mM Tris-HCl pH 8.0, 1 mM EDTA, 1 mM DTT, and 0.1 mg/mL BSA), and allowed to slowly cool to room temperature. The DNA was then end-labeled with gamma 32P-dATP by T4 polynucleotide kinase. In vitro UNG biochemical activity assay was performed as per manufacturers instruction (R&D). In brief, after exposure to UNG2, which deglycosylates uracil, the deglycosylated oligonucleotide was split in half by incubation in Alkali buffer (300mM NaOH, 97% formamide) at 100°C for 10 minutes.
Results
Identification of a novel gene, Ugene, over-expressed in most cancer types
To identify genes potentially involved in colon tumorigenesis, we used GeneChip gene expression microarrays to compare patterns of gene expression in RNA samples extracted from colon cancers versus normal colon epithelium. One transcript, an EST corresponding to GenBank Accession number U46258, was noted to be of particular interest. Specifically, the detectable expression of this transcript was absent in 21 normal colonic mucosal samples (median value 25, corresponding to null expression), but was well detected in 39 malignant colon tumors (median value 125), 43 colon cancer hepatic metastases (median value 107), and 36 colon cancer cell lines (median value 220) (Figure 1A). Ugene induction was found to be specific to colon cancer, and was absent in nine precursor colon adenoma cases (median value 25, corresponding to null expression).
Figure 1. Ugene mRNA expression in normal and cancer samples.
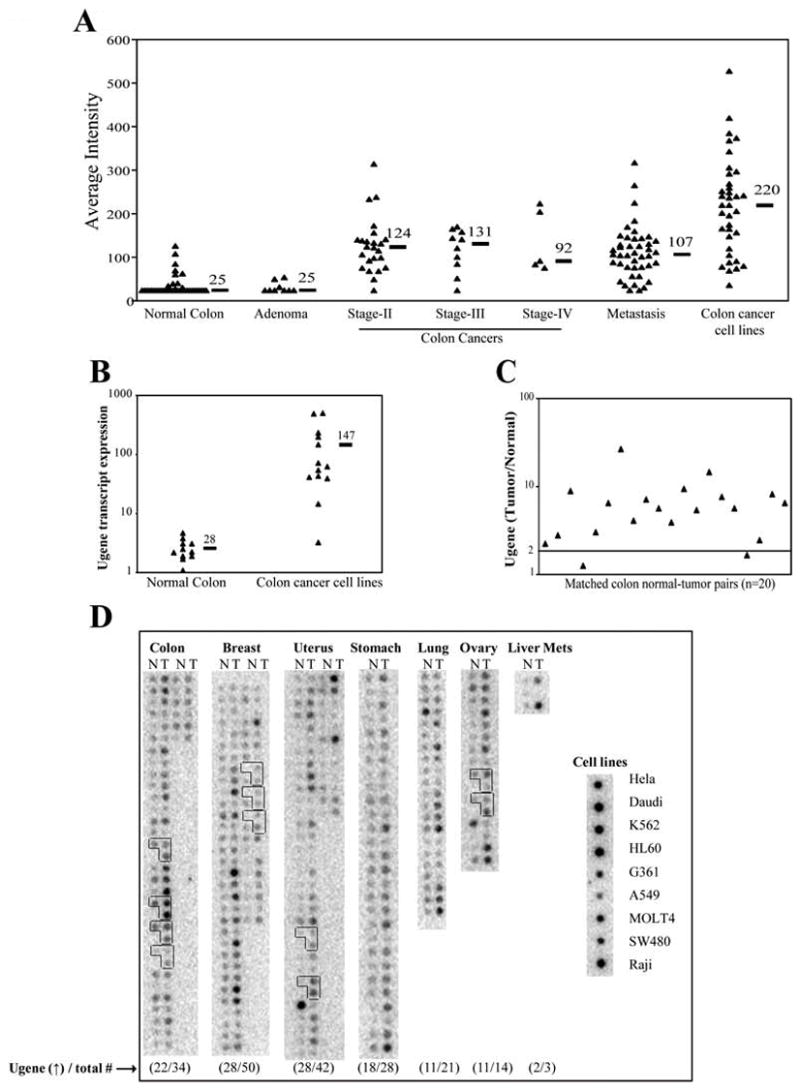
A) Expression of U46258 on GeneChip microarrays. Shown for comparison are analyses of RNA samples from normal colon epithelium, colon adenomas, primary colon cancers of Stages II, III and IV, colon cancer hepatic metastases, and colon cancer cell lines. Horizontal bars denote median expression values within each group. Average intensity (AI) of 25 units corresponds to null expression. B) Real-time PCR measurement of Ugene transcript expression in 11 normal colon epithelial samples versus 13 colon cancer cell lines. Ugene values are normalized against expression of the house-keeping gene, beta-2-microglobulin (B2M). Black bars indicate the mean value for each group. C) The ratio of Ugene expression in colon cancer versus matched normal colon mucosa, as measured by real time PCR in 20 patients. Ugene values are again normalized against B2M. Values represent averages of six replicates. D) cDNA dot blot analysis of Ugene expression level in matched tumor and normal tissue blots from the various organs. N: normal; T: tumor. Numbers below each blot represent the number of tumors with >2-fold increase in Ugene expression relative to their matched normal [Ugene (↑)] versus the number of total samples analyzed (total #). Each L-shaped box of three dots represents normal tissue (left), primary tumor (upper right), and metastases (lower right) from the same patient. Equal sample loading over all lanes were confirmed by rehybriding the blot to a ubiquitin probe (data not shown).
U46258 corresponds to a 123-bp EST sequence mapping to chromosome 1q32. To derive a full length transcript corresponding to this EST, we identified by a BLAST search all ESTs in the dbEST database that mapped to within 20 kb of U46258. Connection RT PCR from colon cancer cell line (SW480 and VACO241) was used to identify ESTs linked to U46258. The 5′ and 3′ rapid amplification of cDNA end (RACE) PCR methods were used to complete the derivation of a colonic transcript that corresponded to U46258 and that contained the complete coding region of a novel gene. This gene, named Ugene, has an mRNA consisting of 4 exons. The open-reading frame encodes a 149 amino acid (aa) protein (16.9kDa) with an ATG start codon in exon 1 and a TAA stop codon in exon 4 (Figure 2). Searching online databases did not identify any known protein motifs contained within the Ugene sequence. BLAST alignment results indicated that the Ugene mRNA sequence shared 99% identity to a predicted human mRNA (accession number XM_001133365), designated as “Homo sapiens similar to family with sequence similarity 72, member A”, which was computationally predicted from the Ugene locus (NCBI contig NT_086602) by the Gnomon gene prediction method.
Figure 2. Structure of the Ugene locus.
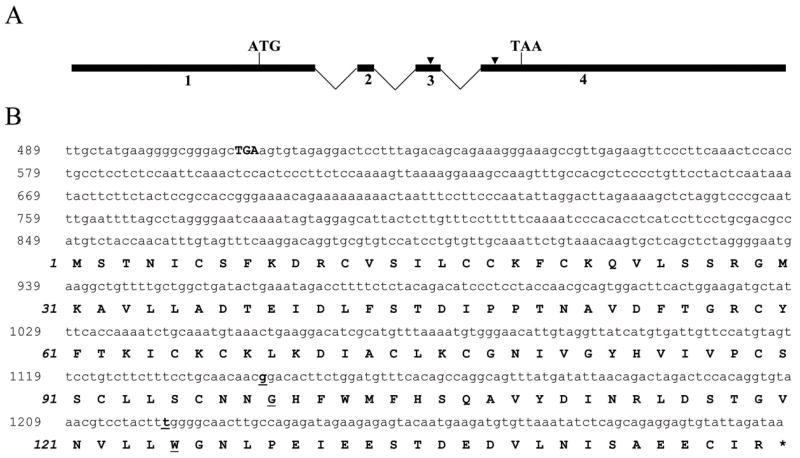
A) Numbered black boxes denote the 4 Ugene exons, with locations of initiator ATG and termination TAA designated. Sites which differentiate Ugene-p and Ugene-q are indicated by arrow heads. B) Nucleotide and deduced amino acid (aa) sequence of complete Ugene (Ugene-p) coding region. The Ugene nucleotide sequence is provided in the upper reading frame and numbered in roman type. The deduced aa sequence is provided underneath the nucleotide sequence, numbered in italic. The in-frame stop codon (TGA) 5′ of the start codon is indicated in boldface. Underlined letters represent codons that differ in Ugene-q.
To further characterize the Ugene transcript, we sequenced 5 or more individually cloned Ugene cDNAs from each of 9 colon cancer cell lines. Surprisingly, each line demonstrated two distinct Ugene transcripts, designated as Ugene-p and Ugene-q, which differed at 3 individual nucleotide positions in exons 1, 3 and 4, respectively. Two of the three nucleotide differences result in amino acid changes at codon 99 (Gly/Arg) and codon 125 (Trp/Arg) (Figure 2). These findings are suggestive that Ugene-p and Ugene-q arise from separate gene loci. In support of this interpretation we additionally demonstrated amplification from genomic DNA of both Ugene-p and Ugene-q specific forms of exons 3 and 4.
BLAST analysis showed 100% correct alignment of the four Ugene-p exons to two different positions on Chromosome 1 (corresponding to GenBank accession numbers NT_086586, NT_086602), suggesting either mis-assembly of Chromosome 1 or multiple copies of this gene. Additionally, BLAST alignment identified 2 exons corresponding to Ugene-q exons 3 and 4 as located on Chromosome 1, but at position different from Ugene-p (corresponding to GenBank accession number NT_034403). Our cDNA cloning supports that in fact four distinct Ugene-q exons exist, likely all located on Chromosome 1. Because Ugene-p is conserved across all mammalian species, we focused on Ugene-p in this study.
As shown in Supplemental Figure 1, Northern blot analysis corroborated that Ugene transcripts are expressed by malignant but not normal colon tissues, detecting a single 2.4 kb Ugene mRNA with moderate to strong intensity in ten of 11 colon cancer cell lines, but in none of 4 normal colon epithelial tissue samples. To provide a more quantitative measurement of Ugene induction, we extended this analysis by employing real-time PCR. Real-time PCR demonstrated only barely detectable Ugene expression in eleven of 11 normal colon epithelial samples (mean value 2.6, range 1.1–4.7), whereas colon cancer cell lines showed an average 56-fold increased level of expression (mean value 147, range 3.3–503), with eleven of 13 colon cancer cell lines showing a greater than 15-fold increase in expression (Figure 1B). To determine expression of Ugene-p versus Ugene-q encoded transcripts, we sequenced individual Ugene cDNA clones from two colon cancer cell lines, SW480 and VACO241. We found that in both cell lines Ugene-p represents 60% of Ugene expression while Ugene-q represents 40% (data not shown).
Over-expression of Ugene in malignant colon cancer was also confirmed by real-time PCR analysis of Ugene mRNA in primary colon cancers versus matched normal colon mucosa from the same individuals. A median increase of 6.8 fold in Ugene expression was observed in cancers versus matched colon normals, with greater than 2-fold increase exhibited by 18 of the 20 tumors (Figure 1C). These 20 colon cancers examined by real-time-PCR constituted a validation set of samples completely independent of those that had been previously characterized on the GeneChip expression microarrays.
Southern blotting of 12 colon cancer cell lines did not demonstrate any increase in Ugene gene copy number as an explanation for Ugene over-expression (data not shown).
To examine whether Ugene might be over-expressed in other cancer types, we probed a cDNA Cancer Profiling Array (BD Biosciences), comparing Ugene expression level in matched tumor and normal tissue from a variety of organs. As expected, a high proportion of colon cancer samples were observed to have elevated expression of Ugene (Figure 1D). Using densitometry, the intensity of the radioactive probe signal from each cDNA sample was quantitated. Twenty-two of 34 (65%) of the colon cancer cases showed greater than 2-fold increased expression of Ugene. Furthermore, we also found Ugene expression elevated in multiple other common cancer types, including breast (56% of cases), lung (52% of cases), stomach (64% of cases), uterus (67% of cases), and ovary (79% of cases).
Ugene encodes a nuclear protein
To investigate the subcellular localization of Ugene encoded protein, a construct expressing V5-epitope tagged Ugene-p protein was transfected into SW480 cells. Figure 3 shows the immunofluorescent staining for the V5 tag (green) in Ugene transfected cells. Results demonstrate that tagged Ugene protein accumulates in nuclei, which were defined by DAPI staining (red). As Ugene is a small protein (16.9kD) and lacks a nuclear localization signal, this accumulation suggested that Ugene might be held in the nucleus through interacting with other nuclear proteins.
Figure 3. Subcellular localization of Ugene.
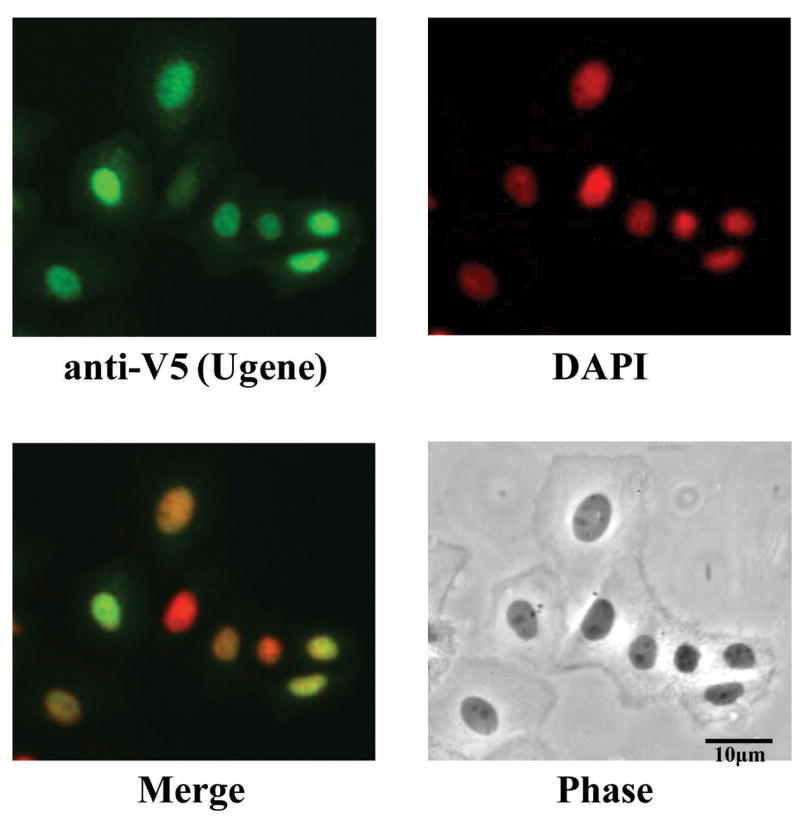
SW480 cells were transfected with constructs expressing V5-tagged Ugene-p protein. The expression of Ugene-p (green) was detected with a V5 antibody and visualized by fluorescent microscopy. Nuclei were stained with DAPI (red).
Ugene binds to UNG2, a base excision DNA repair protein
To look for a potential Ugene partner, we performed a pull-down assay using FLAG-tagged Ugene-p protein over-expressed in SW480 cells. Using mass spectrometry, a protein band that precipitated in the Ugene-p pull-down was identified as uracil DNA glycosylase-2 (UNG2) (data not shown), an enzyme that is involved in base excision repair by catalysis of uracil excision from DNA and is constitutively located in the nucleus (15, 16).
In order to confirm the interaction of Ugene-p and UNG2, we first co-transfected tagged Ugene-p and UNG2 constructs, followed by immunoprecipitation of either protein, and then Western blot analysis of the immunoprecipitates to detect the presence of potential partners. We found Ugene-p and UNG2 co-immunoprecipitated together in assays in which either of the proteins was first pulled down (Figure 4A).
Figure 4. Protein-protein interaction of Ugene-p and UNG2 demonstrated by serial co-immunoprecipitation (Co-IP) Western blot assay.
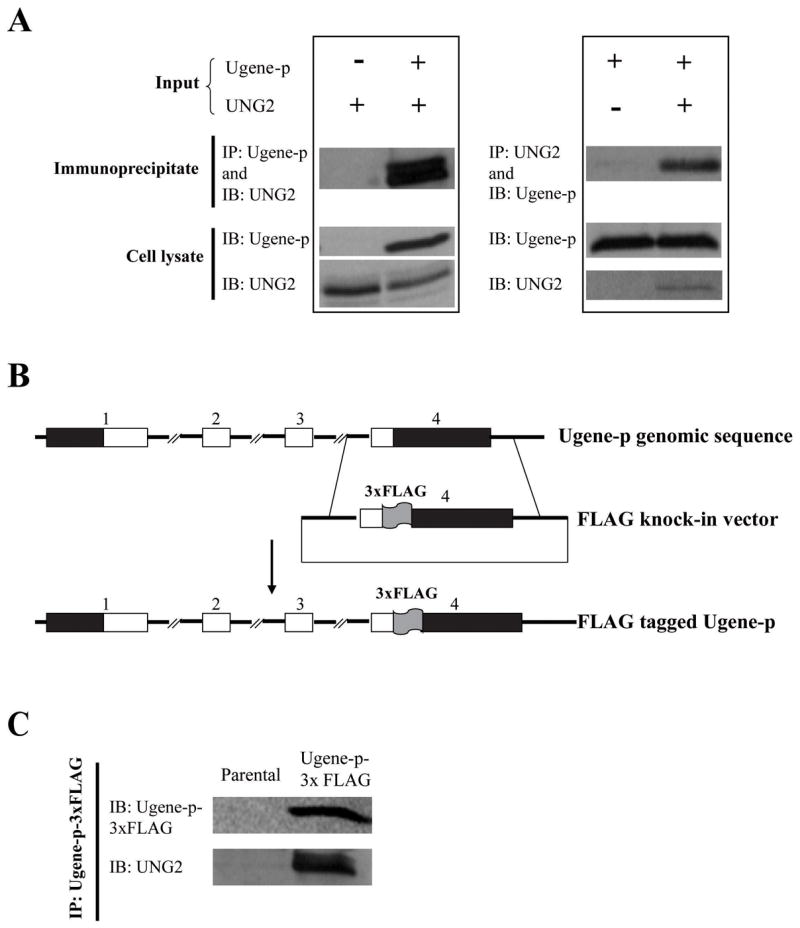
A) The left panel shows HEK 293 cells transfected with an expression vector for UNG2 (V5-epitope-tagged), and cotransfected with an expression vector for Ugene-p (FLAG-epitope-tagged) or the corresponding empty expression vector. Interaction of Ugene-p and UNG2 was tested by immunoprecipitation (IP) of Ugene-p and Western detection (IB) of UNG2 (Upper panel); expression of Ugene-p and UNG2 in the transfected cells is shown by Western blot against epitope tags (lower panel); The right panel shows HEK 293 cells transfected with an expression vector for Ugene-p (V5-epitope-tagged), and cotransfected with an expression vector for UNG2 (FLAG-epitope-tagged) or the corresponding empty expression vector. The upper panel again tests for interaction of Ugene-p and UNG2 by IP of UNG2 and Western detection of Ugene-p; the lower panel shows expression of translated proteins. B) Schematic diagram of the targeting vector for tagging endogenous Ugene-p with 3xFLAG. The numbers above the boxes denote exons; black boxes denote un-translated regions (UTRs); white boxes denote coding sequences. C) Western detection of endogenous Ugene-p and its interaction with endogenous UNG2. Ugene-p was immunoprecipitated from Ugene-p-3xFLAG tagged cells using FLAG antibodies. The upper panel demonstrates endogenous Ugene-p protein detected by Western blot (IB) against the FLAG epitope; the lower panel demonstrates the presence of co-immunoprecipitated UNG2 by Western blot against UNG2. Assay of parental DLD1 cells is shown as controls.
To prove that this binding of Ugene-p and UNG2 was not an artificial result due to protein over-expression, we tagged the endogenous Ugene-p locus in DLD1 cells by somatic cell knock-in of a 3xFLAG epitope at exon 4, corresponding to the COOH-terminus of the protein (Figure 4B) (14). We then co-immunoprecipitated Ugene-p with antibodies against the FLAG-epitope. Western blot analysis confirmed that endogenous UNG2 co-immunoprecipitated with the tagged endogenous Ugene-p protein (Figure 4C). PCR analysis demonstrated no change in total Ugene transcript expression in cells bearing the 3XFLAG knockin epitope and also showed the knockin Ugene allele to be expressed at essentially the same level as the non-targeted alleles (Supplemental Figure 4).
Ugene-p binds to the NH2-terminus of UNG2
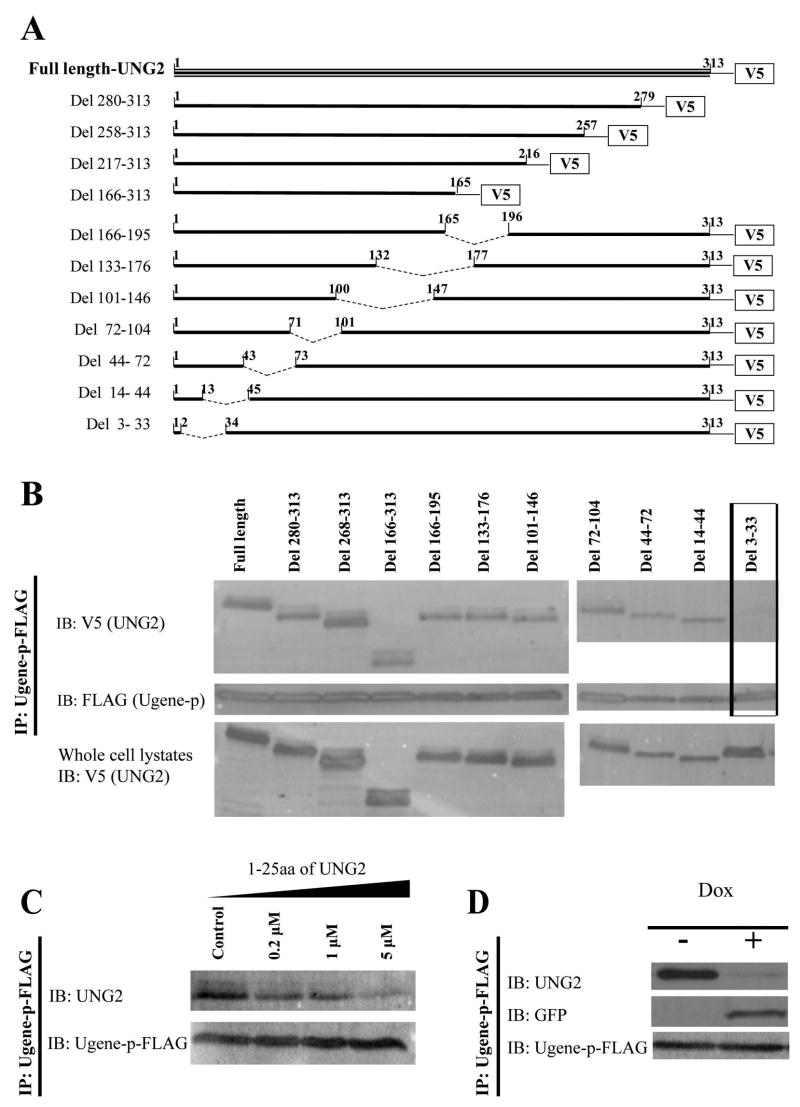
To determine the UNG2 motif responsible for binding to Ugene-p, we made a series of constructs expressing V5-epitope tagged nested UNG2 deletions (Figure 5A). After co-transfecting each of these V5-epitope tagged UNG2 deletion constructs with FLAG-epitope tagged Ugene-p, we immunoprecipitated Ugene-p and performed Western blot analysis to test for co-immunoprecipitation of each of the UNG2 deletion constructs (Figure 5B). One UNG2 deletion that lacked only sequences between codons 3 and 33, showed complete loss of the capacity to bind to Ugene-p. This result suggested that Ugene-p binds to the NH2-terminus of UNG2.
Figure 5. Mapping of the UNG2 domain for binding to Ugene-p.
A) Schematic diagram of constructs expressing nested UNG2 deletions. B) HEK 293T cells were transfected with FLAG tagged Ugene-p and a cDNA encoding either V5-tagged wild type UNG2 or the indicated UNG2 deletion mutants. 48h after transfection, immunoprecipitates prepared by IP with FLAG antibodies were analyzed by Western blot (IB) for UNG2(V5) and Ugene-p(FLAG). Box indicates that deletion of UNG2 amino acid 3-33 abolished the binding to Ugene-p. The lower panel shows expression of V5-tagged wild type UNG2 and UNG2 deletion proteins in whole cell lysates. C) Cell lysates of DLD1 expressing endogenous 3xFLAG-tagged Ugene-p were mixed with artificially synthesized competing peptides (1-25-UNG2, amounts as indicated), then immunoprecipitated with FLAG antibodies. Immunoprecipitates were analyzed by Western blot (IB) for presence of Ugene-p (FLAG) and for co-IP of UNG2 (V5). D) DLD1 cells expressing endogenous 3xFLAG-tagged Ugene-p were transfected with pcDNA6/TR and pcDNA4-1-25-UNG2-GFP, and selected by blasticidine (10μg/ml) and zeocin (200μg/ml) to derive clones conditionally expressing 1-25-UNG2-GFP fusion protein under doxycycline (dox) regulation. These clones are designated as DLD/Ugene-p-3xFLAG/1-25-UNG2-GFP. Interaction of Ugene-p with either UNG2 or the 1-25-UNG2-GFP decoy protein was assayed by IP for Ugene-p with the FLAG antibody, followed by Western blot detection of Ugene-p, GFP, and UNG2, in cells without (dox −) and with (dox +) induced expression of the 1-25-UNG2-GFP decoy protein.
To further test if the UNG2 NH2-terminus is able to bind to Ugene-p, we artificially synthesized peptides encoding the NH2-terminal amino acids 1-25 of UNG2 (1-25-UNG2). We then tested if this peptide could competitively block the binding of endogenous UNG2 to FLAG-epitope tagged endogenous Ugene-p. Figure 5C shows that adding the 1-25-UNG2 peptide into cell lysates blocked UNG2 binding to Ugene-p in a dose dependent fashion. 5μM 1-25-UNG2 competing peptide could compete out almost all Ugene-p binding to UNG2.
To test if the NH2-terminal 1-25aa of UNG2 is sufficient for binding to Ugene-p, we expressed a fusion protein with 1-25-UNG2 fused to green fluorescent protein (1-25-UNG2-GFP) under the regulatory control of doxycycline. We performed this in cells already containing the 3xFLAG-epitope tagged endogenous Ugene-p. Serial immunoprecipitation and Western blot analysis confirmed Ugene-p bound to the (1-25-UNG2-GFP) protein (Figure 5D). Indeed, induction of the 1-25-UNG2-GFP decoy protein could completely out compete and block co-immunoprecipitation of endogenous UNG2 with endogenous Ugene-p (Figure 5D). Therefore, the NH2-terminal 1-25aa of UNG2 are sufficient in vivo for the interaction with Ugene-p.
Interestingly, despite having only 2 amino acids differences, Ugene-q was found not to interact with UNG2 and did not co-immunoprecipitate with it (data not shown). Introducing a single Ugene-q specific codon change, changing tryptophan125 to arginine, was also sufficient to abolish Ugene-p binding to UNG2 (data not shown). Therefore, Trp125 of Ugene-p is required for binding of Ugene-p to UNG2.
Ugene binding does not directly alter UNG2 enzymatic activity or localization
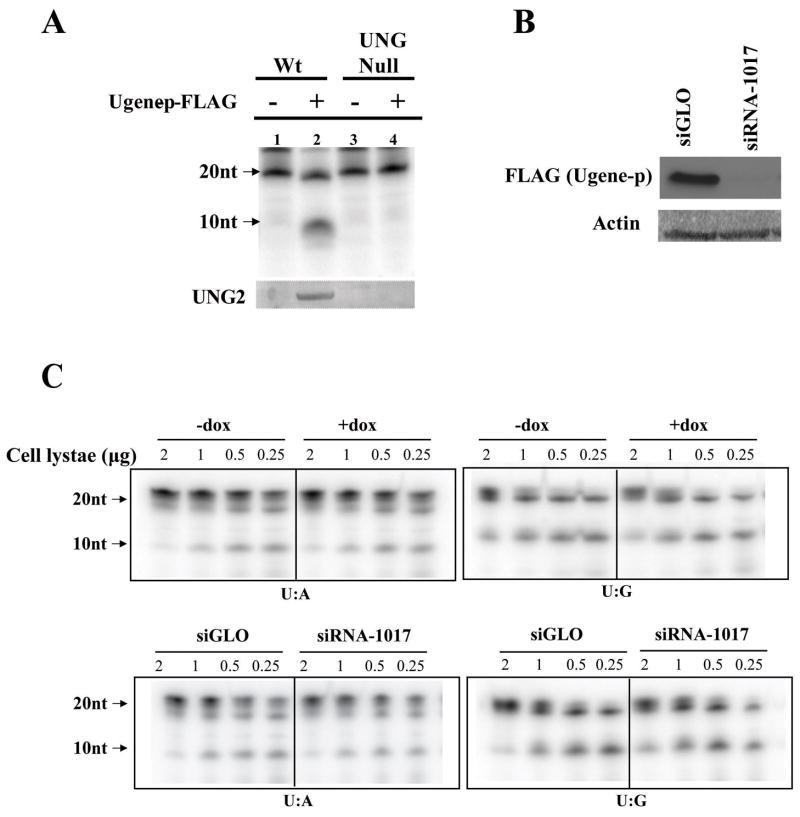
To examine potential functional effects of Ugene-p binding to UNG2, we performed a co-immunoprecipitation to collect UNG2 bound to Ugene-p (pulled down by antibodies against the FLAG-epitope). A biochemical assay showed that UNG2 bound to Ugene-p was an active enzyme, as indicated by initiating a cascade causing cleavage of a uracil containing oligo from the parental 21 nucleotides size down to 10 nucleotides (Figure 6A, lane 2). To ensure the activity in the Ugene-p (FLAG) immunoprecipitates derived from captured UNG2, we repeated the assay in DLD1 cells rendered UNG null by somatic cell knockout (as described in Materials and Methods, Supplemental Figure 3). No activity was detected in Ugene-p immunoprecipitates from UNG null cells. Thus, we conclude that the biochemical activity detected in Ugene-p precipitates from parental DLD1 cells derives from active UNG2 bound to Ugene-p.
Figure 6. Assays of UNG enzymatic activity.
A) Wild type or UNG null DLD1 cells were transfected with a plasmid expressing FLAG tagged Ugene-p or the corresponding empty vector. Cell lysates were then immunoprecipitated with FLAG antibodies. Immunoprecipitates were subjected to a UNG biochemical activity assay, as indicated by presence of a 10nt product (arrow) generated by cleavage of a 21bp input double strand DNA. Input oligos contain a single U:A base pair. The lower panel shows Western blot of UNG2 in Ugene-p immunoprecipitates. B) DLD1 cells expressing endogenous 3xFLAG tagged Ugene-p were transfected with Ugene specific siRNA (siRNA-1017) or control siRNA (siGLO). Ugene-p expression in cell lysates was analyzed by Western blot against the FLAG epitope. C) Using an AAV mediated somatic knockout technique, cells expressing UNG2 only were constructed in the (DLD1/Ugene-p-3xFLAG/1-25-UNG2-GFP) background, The upper two panels show the UNG2 activity in the cell lysates (amount as indicated) without (dox −) and with (dox +) induced expression of the 1-25-UNG2-GFP decoy protein. The lower two panels compare UNG2 activity in lysates (amount as indicated) from cells without (siGLO) and with (siRNA-1017) the suppression of Ugene expression by siRNA. In the left two panels, the input oligos for the assay contain a single U:A base pair; for the right two panels, the input oligos contain a single U:G base pair. UNG2 activity is indicated by autoradioactivity of the 10nt oligos.
To examine whether binding to Ugene-p can alter UNG2 subcellular localization, we expressed V5-epitope tagged UNG2 (UNG2-V5) in the cells conditionally expressing the 1-25-UNG2-GFP fusion protein under doxycycline (dox) regulation. Immunofluorescence against the V5 epitope showed that UNG2 was localized in the nucleus irrespective of expression of the 1-25-UNG2-GFP decoy protein (Supplemental Figure 2). Therefore, expressing a competitor for Ugene-p binding did not alter UNG2 nuclear localization.
The UNG locus encodes both a nuclear protein UNG2, and a mitochondrial isoform UNG1, that both share the same catalytic domain, but are of different sizes (17). In repeated assays, only a UNG2 sized protein was ever detected in Ugene-p immunoprecipitates (data not shown).
To further assay effects of Ugene-p expression on UNG2 activity, we generated cells null for UNG1. This was done by selective knockout of the UNG1 specific exon 1 from the UNG locus. In these cells expressing UNG2 only, we again introduced the 1-25-UNG2-GFP decoy protein under doxycycline regulation. These cells were used to determine UNG2 enzymatic activity under two experimental conditions (Figure 6C). First, we compared UNG2 activity in cell lysates without (dox −) and with (dox +) induced expression of the 1-25-UNG2-GFP decoy protein (upper panel). As shown in Figure 5D, the highly expressed decoy protein totally abolished the interaction of Ugene-p and UNG2, but did not alter UNG2 biochemical activity in the lysates, as shown in Figure 6C (upper panels). Specifically, equal signal intensity of the 10nt cleavage product of the uracil containing oligos was seen in both dox(+) and dox(−) conditions. Second, we compared UNG2 activity in lysates prepared from cells without and with suppression of Ugene expression by siRNA. As demonstrated in Figure 6B, Ugene specific siRNA (siRNA1017) could efficiently suppress Ugene expression by more than 90% at 48 hours after transfection. However, Ugene knock-down did not change the enzymatic activity of UNG2 as shown in Figure 6C (lower panels). These findings were equally true, whether UNG2 activity was analyzed with a 21-bp oligo containing a U:A or a U:G mispair at position 10, which respectively modeled uracil misincorporation into DNA and uracil arising from spontaneous deamination of cytosine. These results suggest that under the experimental condition employed, changing Ugene-p expression did not alter UNG2 biochemical activity.
Discussion
We report here the identification of a novel gene, Ugene-p, whose expression is broadly induced in many human cancer types. Moreover, we demonstrate the interaction of Ugene-p with UNG2, a base excision repair enzyme. The frequent overexpression of Ugene-p in cancer suggests that this gene may participate in the cancer phenotype. A direct assay to test Ugene-p for oncogenic activity, however, revealed no transforming or focus forming activity when tested in epithelial (HMEC) or fibroblast (WI-38, IMR-90, REF-52, NIH 3T3) cells (data not shown).
The interaction of Ugene-p with UNG2, in particular, is highly intriguing, as multiple DNA repair pathways are now recognized as targets for alteration in cancers, including inactivation of genes in the mismatch repair pathway in colon cancers (1) and inactivation of the BRCA1/2 proteins in breast cancers (18). Despite this intriguing association, we have not yet been able to demonstrate a direct regulation of UNG2 repair activity by Ugene-p, in vitro. It is likely however, that the in vivo activity of UNG2 is more complicated than we have been able to model in in vitro assays, as UNG2 in vivo activity involves recognition of misincorporated uracil at the replication fork, and involves recognition of uracils that are spontaneously generated through cytosine deamination in native chromatin, in addition to involving interactions with other members of the BER complex. Suggesting that Ugene-p could promote a specialized function of UNG2, is that immunoprecipitation of over-expressed FLAG-tagged Ugene-p pulled down only a subpopulation of total UNG2 protein (data not shown). Of note, the NH2-terminus of UNG2, to which Ugene-p binds, has been shown to also be bind to the PPM1D phosphatase that dephosphorylates Thr6, effecting a protein modification that is suggested to play an important role in the regulation of UNG2 activity under some circumstances (19). Further analysis of the effect of Ugene-p on UNG2 in these native contexts will be undertaken in future studies.
Genome comparisons show Ugene-p arose as a feature of mammalian cells, in which it is highly conserved, suggesting an important role for the protein in higher organisms. It is, however, unclear from the current genome assemblies whether there are two copies of the Ugene-p on chromosome 1, or whether the chromosome 1 assembly remains in need of revision. In contrast, Ugene-q, which is unable to bind UNG2, is specific to humans, and is absent in other mammals. It is tempting to speculate that Ugene-q may act as a competitor of Ugene-p interactions with other proteins, but testing of this model awaits additional clarification of the functional activities of Ugene-p. Additional future study will also be needed to clarify the role of a second protein, NOSIP, that in our initial pulldown experiment was also identified as binding to Ugene-p, but that on further evaluation proved to not co-precipitate with UNG2 (data not shown).
In summary, we report Ugene-p as a novel gene, commonly over-expressed in human cancers, and participating in a nuclear complex with the base-excision-repair gene, UNG2.
Supplementary Material
Acknowledgments
We thank James D Lutterbaugh for the excellent technical support and Kishore Guda for helpful discussions.
Financial Support: Supported by gift from the Howard Hughes Medical Institute (to S.D.M.) and PHS grant CA116867 and CA120237 (to S.D.M.)
References
- 1.Grady WM, Markowitz SD. Genetic and epigenetic alterations in colon cancer. Annual review of genomics and human genetics. 2002;3:101–28. doi: 10.1146/annurev.genom.3.022502.103043. [DOI] [PubMed] [Google Scholar]
- 2.Li H, Myeroff L, Smiraglia D, et al. SLC5A8, a sodium transporter, is a tumor suppressor gene silenced by methylation in human colon aberrant crypt foci and cancers. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:8412–7. doi: 10.1073/pnas.1430846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moinova HR, Chen WD, Shen L, et al. HLTF gene silencing in human colon cancer. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:4562–7. doi: 10.1073/pnas.062459899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan M, Rerko RM, Platzer P, et al. 15-Hydroxyprostaglandin dehydrogenase, a COX-2 oncogene antagonist, is a TGF-beta-induced suppressor of human gastrointestinal cancers. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:17468–73. doi: 10.1073/pnas.0406142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang Y, Siegel PM, Shu W, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer cell. 2003;3:537–49. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 6.Xin B, Platzer P, Fink SP, et al. Colon cancer secreted protein-2 (CCSP-2), a novel candidate serological marker of colon neoplasia. Oncogene. 2005;24:724–31. doi: 10.1038/sj.onc.1208134. [DOI] [PubMed] [Google Scholar]
- 7.Willson JK, Bittner GN, Oberley TD, Meisner LF, Weese JL. Cell culture of human colon adenomas and carcinomas. Cancer research. 1987;47:2704–13. [PubMed] [Google Scholar]
- 8.Markowitz S, Wang J, Myeroff L, et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–8. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 9.Platzer P, Upender MB, Wilson K, et al. Silence of chromosomal amplifications in colon cancer. Cancer research. 2002;62:1134–8. [PubMed] [Google Scholar]
- 10.Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nature genetics. 1999;21:20–4. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- 11.Brunschwig EB, Wilson K, Mack D, et al. PMEPA1, a transforming growth factor-beta-induced marker of terminal colonocyte differentiation whose expression is maintained in primary and metastatic colon cancer. Cancer research. 2003;63:1568–75. [PubMed] [Google Scholar]
- 12.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods in molecular biology. 2000;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 13.Kohli M, Rago C, Lengauer C, Kinzler KW, Vogelstein B. Facile methods for generating human somatic cell gene knockouts using recombinant adeno-associated viruses. Nucleic acids research. 2004;32:e3. doi: 10.1093/nar/gnh009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Guo C, Chen Y, et al. Epitope tagging of endogenous proteins for genome-wide ChIP-chip studies. Nature methods. 2008;5:163–5. doi: 10.1038/nmeth1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kavli B, Sundheim O, Akbari M, et al. hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. The Journal of biological chemistry. 2002;277:39926–36. doi: 10.1074/jbc.M207107200. [DOI] [PubMed] [Google Scholar]
- 16.Nilsen H, Haushalter KA, Robins P, Barnes DE, Verdine GL, Lindahl T. Excision of deaminated cytosine from the vertebrate genome: role of the SMUG1 uracil-DNA glycosylase. The EMBO journal. 2001;20:4278–86. doi: 10.1093/emboj/20.15.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nilsen H, Otterlei M, Haug T, et al. Nuclear and mitochondrial uracil-DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene. Nucleic acids research. 1997;25:750–5. doi: 10.1093/nar/25.4.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fackenthal JD, Olopade OI. Breast cancer risk associated with BRCA1 and BRCA2 in diverse populations. Nature reviews. 2007;7:937–48. doi: 10.1038/nrc2054. [DOI] [PubMed] [Google Scholar]
- 19.Lu X, Bocangel D, Nannenga B, Yamaguchi H, Appella E, Donehower LA. The p53-induced oncogenic phosphatase PPM1D interacts with uracil DNA glycosylase and suppresses base excision repair. Molecular cell. 2004;15:621–34. doi: 10.1016/j.molcel.2004.08.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.