Figure 1.
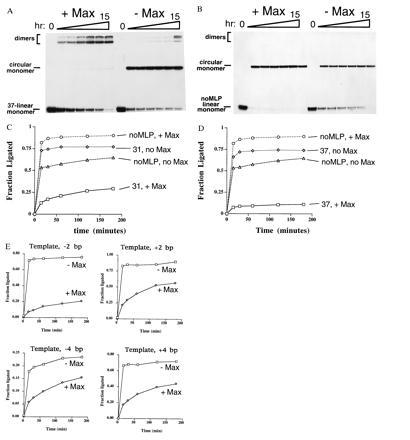
Inhibition of circularization by Max. Ligation kinetic studies were carried out using 156-bp linear DNAs containing the CACGTG (derived from adenovirus major late promoter, MLP) Max recognition sequence either in phase (31 bp) or out of phase (37 bp) relative to dA tracts within the same fragment. Fragments lacking the binding site are indicated as noMLP. Incubations were ended at 0, 15 min, 30 min, 1 hr, 2 hr, 3 hr, and 15 hr. (A) Electrophoretic profile of typical ligation reactions for fragments containing the Max binding site. Higher-order forms, including linear or circular dimers are indicated at the top of the gel. (B) Ligation reactions for fragments lacking the Max binding site (noMLP linear monomer). (C) Phase-independent inhibition of circularization. Ligation products were quantified by PhosphorImager and the molar fraction present as circular monomer was plotted as a function of time. Probes either contained the CACGTG binding site for Max at a spacing of 31 bp (in phase) from the center of the last dA tract (indicated as 31) or lacked the CACGTG binding site (indicated as noMLP), and ligations were run either in the presence or absence of purified Max protein as indicated. (D) Ligation conditions identical to C except that the CACGTG-containing template was spaced 37 bp (out of phase) from the center of the adjacent A tract (indicated as 37). DNA templates lacking the CACGTG binding site are indicated as noMLP. (E) Control for twist in circularization kinetics. Linear templates were produced exactly as above, but varying in length −2, −4, +2, or +4 bp relative to the 156-bp 31 linear template. Ligation kinetics were carried out as above, containing or lacking Max as indicated. In each case, the presence of Max impeded circularization, suggesting that twist does not explain the lack of circularization of the 156-bp templates.
