3.
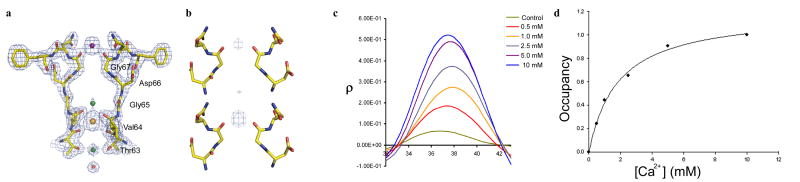
Ca2+ binding in the NaK selectivity filter. (a) 2Fo-Fc ion omit map (1.5σ) of the NaKNΔ19-Na+ complex co-crystallized with 10mM CaCl2, showing electron density for a Ca2+ ion (purple sphere) at the external entrance. Na+ and the contaminant at site 3 are modeled as green and orange spheres, respectively. Electron density for the protein backbone is also shown as a blue mesh. (b) F0.5mM Ca − F0mM Ca (upper) and F5mM Ca2+ − F0mM Ca2+ (lower) difference map contoured at 10 σ showing increasing intensity of Ca2+ binding upon increasing Ca2+ concentration. Dotted lines indicate the position of central axis along which the 1-D electron density was calculated. (c) 1-D electron density profile of Ca2+ binding at the external entrance showing increased Ca2+ binding at increasing concentrations. The control is from a difference map between two native crystals grown in the absence of Ca2+. (d) Occupancy of Ca2+ binding at the external site was calculated by integration of the peaks in 1-D electron density profile and normalizing them against the 10 mM Ca2+ peak. The plot of occupancy against Ca2+ concentration give rise to a Kd value of about 1.8 mM upon fitting the curve to a single component Langmuir equation.