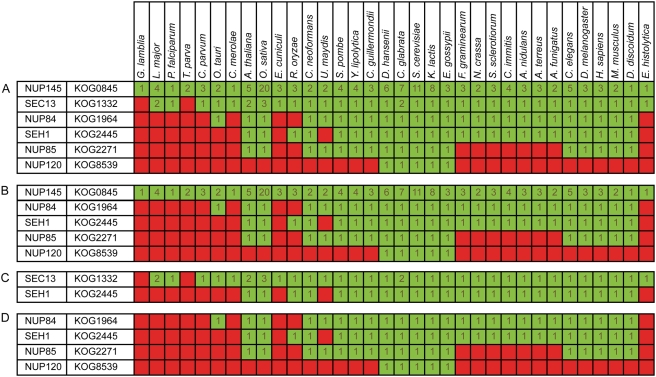
Figure 1. Example of a flexibly evolving complex: Nup84 subcomplex of the nuclear pore complex.
(A) The profile of the Nup84 complex, red indicating absence, green presence (number of paralogs in dark green). The raw score of this complex is (5,0), which means that there are 5 species in which this complex is completely present and none in which this complex is completely absent. The cohesiveness score, which is the fraction of random modules of the same size which score better both in the number of species in which the module is present as well as in the number of species in which the module is absent, is 0.48. This complex from the Aloy dataset occurs also in the MIPS dataset and, with some additional subunits, in the PE and Socio-affinity clusters, so it passes the cross-comparison filter without losing any subunits. (B) The profile after cross-comparison with TAP data. SEC13, which is also part of the COPII complex, has the lowest PE score with the other subunits and has a higher propensity to interact with a protein outside the module (namely with SEC31, an other member of the COPII complex) than with any other member of this module. Removal of this protein from the module results in a subcomplex which is not evolving more cohesively than the original module. (C) Apart from improving the module definition, we attempt to filter possible noise originating from the use of orthologous groups to describe a modules evolutionary dynamics. KOG0845, KOG1964, KOG2271 and KOG8539 are considered unreliable because they have less than 90% overlap with a orthoMCL derived orthologous group. Removal of those orthologous groups leads to a more cohesively evolving module, with a raw score (24,2) and a cohesiveness score 0.87. (D) Removal of orthologous groups which are likely to have functionally differentiated (groups containing many inparalogs, in this example KOG0845 and KOG1332) results in a submodule which we consider evolutionary cohesive: it has a raw score of (5,8) and a cohesiveness score of 0.996. More details on this module and some additional examples can be found in Text S2.