Figure 2. Schematic of the Random Mutation Capture assay.
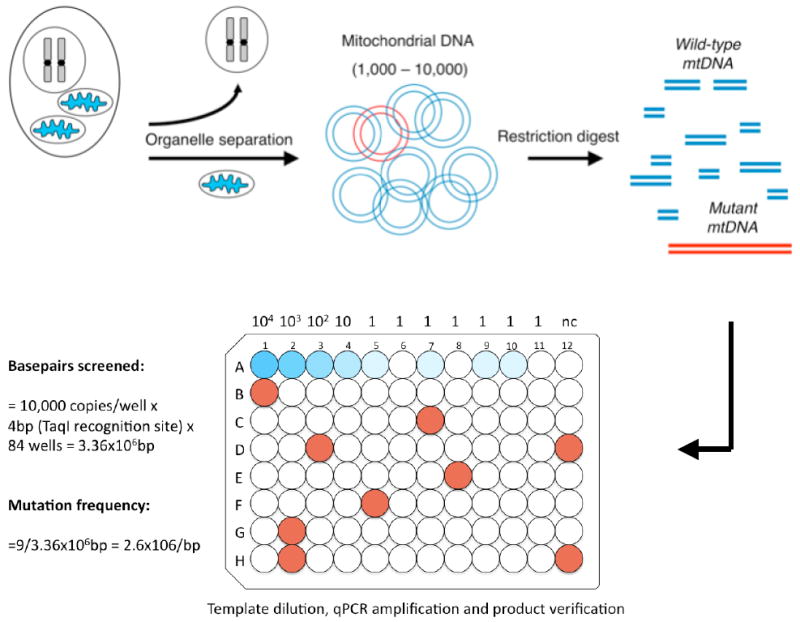
The RMC-assay consists of 4 steps, organelle separation, DNA extraction, DNA digestion, and qPCR amplification. In order to calculate the mutation frequency, mtDNA is displayed in a 96-well format. In the example presented above, 10,000 molecules of the mitochondrial genome are inserted into rows B-H. In each of these wells, a PCR reaction is attempted across the TaqI restriction site. Here, 9 wells, displayed in red contained an amplified molecule. Sequencing of each of these PCR reactions confirmed that a mutation was present in the TaqI restriction site. Serial dilutions, from 10,000 copies to 1, in row A, are used to confirm the copy number present in each well, and provide an important control for PCR efficiency. At an estimated 1 copy per well (wells A5-A11), some wells do, and some do not contain an amplified DNA molecule. The mutation frequency can be calculated as follows; If 10,000 copies of mtDNA are screened per well, that is equivalent to a screen of 40,000 basepairs per well, since the TaqI site is 4bps long, and a mutation in any one of these basepairs will render it resistant to cleavage. 84 wells were screened in this experiment, which amounts to 3.36×106bps screened. 9 Mutants were found, yielding a mutation frequency of 2.6×10-6.
