Figure 2.
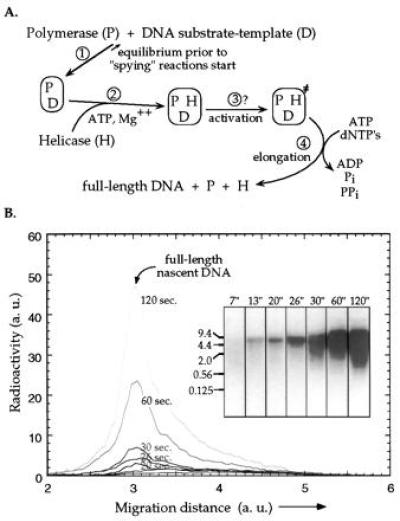
Strand-displacement DNA synthesis on the long dsDNA helicase–polymerase substrate-template construct catalyzed by gp41 and gp43. (A) Reaction pathway for the coupled helicase–polymerase assay. The binding of DNA polymerase to the DNA construct (step 1) is an equilibrium process in which association is greatly favored. The helicase and polymerase reactions are then triggered by the loading of the ATP-activated helicase (step 2). The fully assembled ternary complex unwinds the dsDNA template and synthesizes a new DNA strand by elongating the existing primer. An activation step (step 3) may or may not be involved after the assembly of the ternary complex and before the two-protein replication system enters the elongation phase (see Discussion). The lag-time in full-length DNA production corresponds to the total time required for the activation and elongation reactions (steps 3 and 4). (B) Analysis of the products of the coupled assay by DNA strand-separating alkaline agarose gel electrophoresis. Reactions were performed in the presence of 7.5% PEG and a trace amount of [α-32P]ATP. The resulting radioactively-labeled DNA strands were analyzed by alkaline agarose gel (1%) electrophoresis (Inset) and detected using the AMBIS radioactivity scanning system. The numbers at the top of the lanes denote reaction time (in seconds) while those at the left side of the gel indicate the positions to which various DNA fragments have migrated. The scans represent the distribution of radioactivity along the lanes of the gel. Both the amount of radioactivity and the migration distance are in arbitrary units. Note that even though the images in the gel show significant broadening of the DNA bands at longer times, the scans (which have a much higher dynamic range) indicate little change in the relative distributions of fragments within the lanes.
