Abstract
Immunomics research uses in silico epitope prediction, as well as in vivo and in vitro approaches. We inoculated BALB/c (H2d) mice with 17DD yellow fever vaccine to investigate the correlations between approaches used for epitope discovery: ELISPOT assays, binding assays, and prediction software. Our results showed a good agreement between ELISPOT and binding assays, which seemed to correlate with the protein immunogenicity. PREDBALB/c prediction software partially agreed with the ELISPOT and binding assay results, but presented low specificity. The use of prediction software to exclude peptides containing no epitopes, followed by high throughput screening of the remaining peptides by ELISPOT, and the use of MHC-biding assays to characterize the MHC restrictions demonstrated to be an efficient strategy. The results allowed the characterization of 2 MHC class I and 17 class II epitopes in the envelope protein of the YF virus in BALB/c (H2d) mice.
Abbreviations: ELISPOT, Enzyme-linked immunosorbent spot assay; IC50, 50% inhibitory concentration; YF, yellow fever; DC, dendritic cells; SFC, spot-forming cells.
Keywords: Epitope, Yellow fever virus, Immunome, ELISPOT, Binding assay, Prediction software
Introduction
In the last decade, many groups have devoted attention to the study of the immunomes of a variety of viruses with the goal of obtaining new insights that could support the development and improvement of vaccines, viral diagnosis, and further our understanding of the immune system (Peters et al., 2005, Sette and Fikes, 2003, Sette et al., 2005).
Immunome, as defined by Sette and co-workers (Sette et al., 2005), is the detailed map of immune reactions of a given host interacting with a foreign antigen, and immunomics can be defined as the study of immunomes. The identification of the T and B cell immune response targets allows the design of antigenic formulations focused on selected epitopes, for instance, promoting more robust responses against subdominant epitopes contained in conserved sequences that do not easily tolerate mutants escape. In addition, it makes it possible to avoid the presence of inhibitory epitopes (Disis et al., 1996), allows the combination of many epitopes from different proteins or organisms (Sercarz et al., 1993) and the avoidance of immunopathogenic epitopes. Hence, immunomics offers a powerful approach to the rational design of vaccines (Sette and Fikes, 2003).
In addition to prediction software approaches (Moutaftsi et al., 2006, Yewdell, 2006), ELISPOT (Anthony and Lehmann, 2003), flow cytometry (Hoffmeister et al., 2003), binding assays (Sidney et al., 1998), and human leukocyte antigen (HLA)-transgenic mice (Sette and Fikes, 2003) are the most frequently used tools for epitope discovery. Among those, ELISPOT offers many positive advantages with regards to characterizing the immune response (e.g. looking at different cytokines and/or subpopulations of cells), by offering some degree of automation, broader options for readouts (e.g., IFN-γ, IL-2, and IL-4), the screening of large number of peptides, and rapid results (Anthony and Lehmann, 2003). Although the length of the peptides used in ELISPOT assays can vary from 9 to 20 amino acids, libraries of 15 amino acids (15mers) with an offset of 11 amino acids seem to represent the best compromise between epitope coverage of a protein sequence and the ability to detect CD4+ and CD8+ responses (Kiecker et al., 2004).
Several factors are known to modulate the repertoire of T cells responses, such as the protein's three-dimensional structure, its processing, presentation, MHC–peptide affinity, T cell receptor (TCR) diversity, and avidity. These factors can be highly affected by how the antigen formulations are prepared and presented to the host organism. Therefore, a high-quality characterization of an immunome must include the testing of multiple immunization protocols using antigens inoculated via different routes and doses, with the immune responses tested at different points in time (Assarsson et al., 2007). Since it is nearly impossible to test all the potential antigen formulations and screen all the peptides that might induce responses, it is important to include in the immunome study reductionist approaches that are subjected to fewer variables. One of the most commonly used approaches is the biochemical MHC–peptide binding assay. The direct in vitro evaluation of peptides that can bind MHC molecules, as determined by competition assays, is likely to identify a larger pool of possible epitopes that would not actually bind in vivo, due to various processing and chaperone mechanisms, which are part of the immunodominant shaping mechanisms (Yewdell, 2006).
The use of prediction software has also become a very important tool for epitope discovery. Such software is intended to save time and money by allowing investigators to pre-screen the sequence of a protein or a list of peptides for the presence of possible targets for the immune response. Different mathematical models are used in the search for peptides that can bind to MHC molecules with high affinity. These models can be as simple as matrices of frequencies of individual amino acids in a given position in a peptide sequence or as complex as artificial neural network models; this topic has been reviewed elsewhere (Braga-Neto and Marques, 2006).
The YF virus is an interesting target for immunome studies, with a number of useful relevant tools available that facilitate these studies, such as an effective human live-attenuated virus vaccine and accepted animal challenge models. YF is a deadly arthropod-borne virus of the flavivirus family that is currently circulating in tropical and subtropical areas of the world (Figueiredo, 2007, Monath, 2001, World Health Organization, 2003). The 17D, 17D-213, and 17DD, YF strains used for human vaccination are products of multiple passages of the virulent parental Asibi virus (Galler et al., 1997). The molecular basis of the attenuation has been identified, and all the vaccine strains have been shown to differ from the wild-type virus in a few amino acids (dos Santos et al., 1995, Galler et al., 1997, Jennings et al., 1993, Post et al., 1992). The strain 17D has been successfully used worldwide for more than 65 years. It has an extraordinary record of safety, conferring long-lasting immunity for up to 35 years in as many as 99% of those vaccinated (Poland et al., 1981, World Health Organization, 2003).
The humoral neutralizing response against the YF envelope protein, one of its three structural proteins, has been recognized as one of the major mechanisms to account for the protection elicited by the vaccine (Monath, 1986, Monath and Barrett, 2003), although data increasingly point to an important role for the cellular arm of the immune response in protection against infection (Co et al., 2002). A few MHC-restricted T cell epitopes have been described in mice (Regner et al., 2001, van der Most et al., 2002) and humans (Co et al., 2002), but their contribution to the immunity induced by the vaccine is still not understood. It was recently reported that the YF 17D vaccine can infect dendritic cells (DC) (Barba-Spaeth et al., 2005) and is able to activate innate immune responses through toll-like receptors (TLR) (Querec et al., 2006).
We used the 17DD YF virus as a surrogate model for our immunomics studies of the YF virus structural proteins. Murine T cell responses were evaluated ex vivo through ELISPOT IFN-γ assays with overlapping peptide libraries encompassing the entire length of each of the three YF structural proteins (envelope, capsid, and membrane). These results were then compared to a computational T cell epitope prediction method, PREDBALB/c, which is based on binding matrix models for each of the five MHC alleles present in the BALB/c mouse, and to binding assay results for all five H2-D alleles. All the epitopes described in this study can be found in the Immune Epitope Database —IEDB (http://www.immuneepitope.org/home.do) (Peters et al., 2005).
Results
Evaluation of the kinetics and dose–response of BALB/c mice to the immunization with the human 17DD YF vaccine
In order to define a suitable immunization protocol for immunome studies using the human 17DD YF virus, groups of BALB/c mice were immunized three times, at 2-week intervals, with either a high (105 pfu) or low dose (104 pfu) of the 17DD YF vaccine and were assayed ex vivo by IFN-γ ELISPOT assay using 16mer peptides from the YF envelope protein, (Supplemental Table 1). A detectable T cell response was observed in both mice groups with the high and low doses 14 days after the first immunization (Supplemental Fig. 1A and B). The second immunization led to a more focused T cell response to fewer 16mer peptides (Supplemental Figs. 1C and D) and the third immunization did not change the pattern observed after the second immunization (Supplemental Fig. 1E and F). The evaluation of the humoral response after each immunization (data not shown) indicated that after two immunizations with either the high or low dose of the vaccine, both groups reached similar levels of IgG anti-YF (ELISA with virus lysate), and that the third immunization did not enhance the antibody levels. The results of these experiments (Supplemental Fig. 1) indicated that an immunization protocol with two low dosed of the 17DD YF vaccine given three weeks apart would be enough to induce an optimal T cell response, and hence, such protocol was applied in the subsequent experiments, except when indicated.
Analysis of CD4+ and CD8+ responses to the YF envelope protein peptides
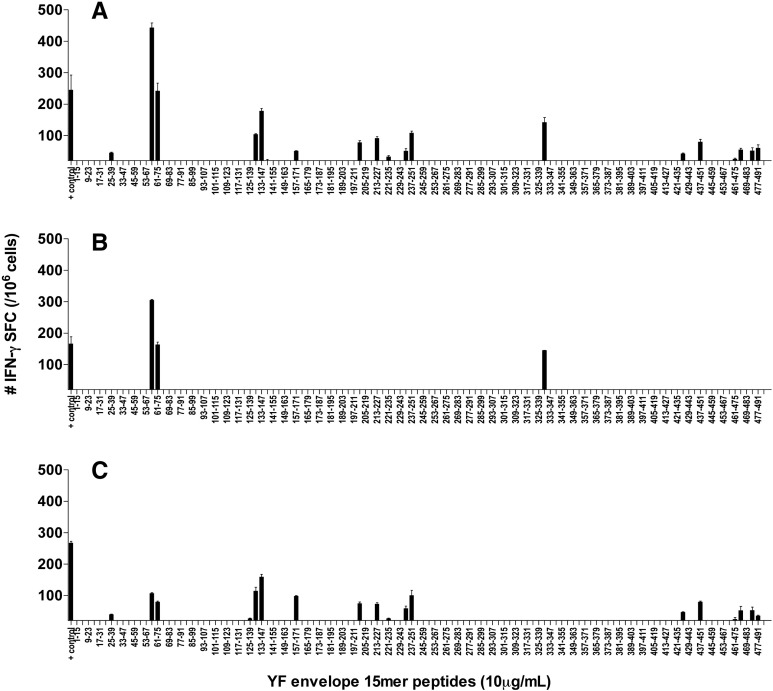
Seven days after the second immunization, the splenocytes of the mice receiving two low doses (104 pfu) of the human 17DD YF vaccine 3 weeks apart were assayed by ELISPOT with 15mer peptides (15 × 11) of the 17DD YF envelope protein (Supplemental Table 2). A high number of IFN-γ SFC was observed against several peptides of the envelope protein (Fig. 1A) and the main positive responses were directed to the same regions in the YF envelope protein previously observed with the 16mer peptides (16 × 10) (Supplemental Fig. 1).
Fig. 1.
Response of total splenocytes and CD4+ and CD8+ T cells to the peptides of the YF envelope protein. BALB/c mice were immunized on day 0 and boosted on day 21 with 104 pfu of the human 17DD YF vaccine, and the splenocytes were tested in IFN-γ ELISPOT assays 7–10 days after the boost. The peptides used for in vitro stimulation were 15mers, overlapping by 11 amino acids and comprising the length of the envelope protein (10 μg/mL). (A) splenocytes; (B) CD4-depleted splenocytes; (C) CD8-depleted splenocytes. Figures represent the average of two to four experiments performed with a pool of three to five mice each. Bars indicate the mean ± SD.
We further characterized the cellular response using splenocytes depleted of CD4+ or CD8+ lymphocytes. The depletion approach consistently yielded > 95% of depletion, as judged by flow cytometry (data not shown). CD4-depleted cells, which were used to address the CD8+ response, showed significant numbers of IFN-γ SFC in response to three of the peptides (Fig. 1B), while the CD8-depleted cells, which correspond to the CD4+ response, showed significant numbers of IFN-γ SFC in response to 17 peptides (Fig. 1C). These results were compatible with the findings obtained with total splenocytes (Fig. 1A) and characterized the immunogenic peptides either as MHC class I or class II antigens. As an additional control, experiments were performed with purified CD4+ or CD8+ T cells incubated with splenocytes from naive mice as APC, and the ELISPOT results were comparable to those obtained with depleted populations (data not shown). Two of the peptides (E57–71 and E61–75) promoted IFN-γ secretion from both CD4+ and CD8+ T cells, suggesting that these 15mers contain epitope motifs for class I and class II molecules, whereas the remaining immunogenic peptides induced either CD4+ or CD8+ responses.
The T cell repertoire of the immunized mice was also investigated at 4.5 and 9 months after the initial immunization protocols, with or without a recall boost right before the experiments (Supplemental Fig. 2). In these long-term memory experiments, we observed the same set of immunogenic peptides that was found seven days after the second 17DD YF immunization. The peptides corresponding to the CD8+ response appeared to be the most dominant immune responses present at all times tested.
Characterization of the CD8+ responses to the YF envelope protein peptides
The CD8+ T cells preferentially recognize peptides of 8–10 amino acids present in the groove of MHC class I molecules. Therefore, the pattern of CD8+ response to 15mers does not define the minimal sequences that are being recognized. To address this issue, we examined how CD8+ T cells from mice immunized with our standard 17DD YF virus protocol react in vitro with all the possible 9mers that are present within each of the three CD8-immunogenic 15mer peptides (Table 1 ; Supplemental Table 5). For the first two CD8-immunogenic 15mers, E57–71 and E61–75, we considered the amino acid sequence between position E57 and E75 of the envelope protein, which results in 11 peptides of 9 amino acids overlapped by 8 amino acids (9 × 8). For the third CD8-immunogenic 15mer (E329–343), the sequence between E329 and E343 was considered, i.e., 7 peptides of 9 amino acids, overlapped by 8 amino acids.
Table 1.
Results of ELISPOT, binding assay and PREDBALB/c prediction for the 9mer peptides from the CD8-positive sequences of the yellow fever envelope protein
| 9mer position (Yellow Fever Envelope) |
Sequence |
# IFN-γ SFC (CD4-depleted splenocytes/106) (2) | Binding assay (IC50 in nmol) (PREDBALB/c score) |
||
|---|---|---|---|---|---|
| E57 to E75 | RKVCYNAVLTHVKINDKCP(1) | Dd (3) | Kd (3) | Ld (3) | |
| 57–65 | RKVCYNAVL(4) | 1 | > 70,000(5) | > 70,000 | 39,442 |
| (7.98) | (5.72) | (4.70) | |||
| 58–66 | KVCYNAVLT | 93 | > 70,000 | > 70,000 | > 70,000 |
| (2.32) | (1.84) | (0.90) | |||
| 59–67 | VCYNAVLTH | 265 | > 70,000 | 211⁎ | 55,368 |
| (7.98) | (1.60) | (4.62) | |||
| 60–68 | CYNAVLTHV | 403 | > 70,000 | 1.0⁎ | > 70,000 |
| (6.48) | (8.66⁎⁎) | (5.18) | |||
| 61–69 | YNAVLTHVK | 143 | > 70,000 | 93⁎ | > 70,000 |
| (3.46) | (1.92) | (0.90) | |||
| 62–70 | NAVLTHVKI | 7 | > 70,000 | 1542 | > 70,000 |
| (8.42⁎⁎) | (5.56) | (0.30) | |||
| 63–71 | AVLTHVKIN | 60 | > 70,000 | 54,670 | 20,708 |
| (4.10) | (1.10) | (0.88) | |||
| 64–72 | VLTHVKIND | 60 | > 70,000 | > 70,000 | > 70,000 |
| (2.94) | (5.22) | (2.22) | |||
| 65–73 | LTHVKINDK | 51 | > 70,000 | > 70,000 | > 70,000 |
| (4.02) | (1.60) | (1.50) | |||
| 66–74 | THVKINDKC | 1 | > 70,000 | > 70,000 | > 70,000 |
| (0.80) | (2.26) | (0.40) | |||
| 67–75 | HVKINDKCP | 6 | > 70,000 | > 70,000 | 48,666 |
| (1.64) | (0.58) | (2.5) | |||
| E329 to E343 | PCRIPVIVADDLTAA(6) | ||||
| 329–337 | PCRIPVIVA | 158 | > 70,000 | > 70,000 | > 70,000 |
| (4.78) | (0.50) | (0.44) | |||
| 330–338 | CRIPVIVAD | 371 | > 70,000 | 915 | 54,239 |
| (2.36) | (2.40) | (2.12) | |||
| 331–339 | RIPVIVADD | 140 | > 70,000 | > 70,000 | > 70,000 |
| (8.50⁎⁎) | (4.18) | (1.42) | |||
| 332–340 | IPVIVADDL | 63 | > 70,000 | 33,005 | 56⁎ |
| (6.72) | (6.00) | (7.50) | |||
| 333–341 | PVIVADDLT | 26 | > 70,000 | > 70,000 | 3719 |
| (3.40) | (1.80) | (0.00) | |||
| 334–342 | VIVADDLTA | 86 | > 70,000 | > 70,000 | 19,242 |
| (2.70) | (3.10) | (1.50) | |||
| 335–343 | IVADDLTAA | 46 | > 70,000 | > 70,000 | 38,032 |
| (3.46) | (1.96) | (0.90) | |||
(1) Sequence of 19 amino acids that correspond to 15mer peptides E57–71 and E61–75 that were positive for CD8 response by ELISPOT (Fig. 2B); (2) peptides were considered ELISPOT positives when > 20 SFC/106 cells; (3) molecules of H2d class I; (4) sequence of each 9mer in relation to the sequence from where it is found in the envelope protein; (5) IC50 for the binding assay for class I molecule (nmol); (6) sequence of 15 amino acids from the 15mer peptide E329–343 (Fig. 2B); ⁎values of IC50 below the cutoff of 500 nmol, suggestive of a possible binder peptide; ⁎⁎ score above 8 from PREDBALB/c software prediction.
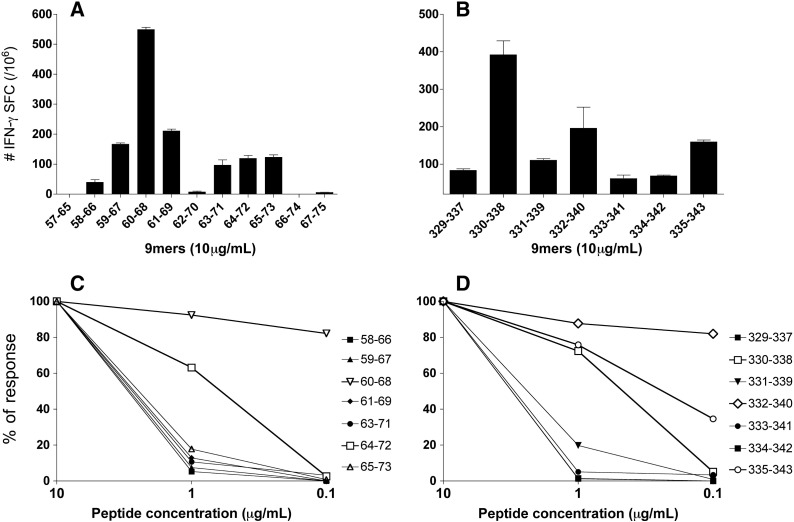
The results of the IFN-γ ELISPOT assays using 9mer peptides to stimulate the in vitro responses are presented in Figs. 2A and B. Within the first set of 9mers (E57 to E75), the 9mer peptide E60–68 induced the highest number of IFN-γ SFC, while in the second set of 9mers (E329–343), the peptide E330–338 stimulated the highest number of IFN-γ SFC. However, in both 9mer sets, other 9mer peptides were also able to stimulate a significant number of IFN-gamma SFC. In order to determine how the concentration of peptide used in the ELISPOT assay could influence the results, we conducted a second round of experiments using decreasing in vitro concentrations of peptide (10, 1, and 0.1 μg/mL) (Figs. 2C and D). In these experiments, peptide E60–68 induced the highest number of IFN-γ SFC at all concentrations tested, showing a high in vitro stimulatory activity for CD8+ T cells of mice immunized with the 17DD YF virus (Fig. 2C). Peptide E64–73 showed an intermediate stimulatory level, while all the other peptides in the same set induced measurable IFN-γ secretion only at the highest concentration of 10 μg/mL.
Fig. 2.
CD8 responses to 9mer peptides of the YF envelope protein. Splenocytes of BALB/c mice immunized twice with the human 17DD YF vaccine were used for IFN-γ ELISPOT assays. The percentage of responses for each peptide was calculated according to the formula: number of spots at 1 μg/mL (or 0.1 μg/mL) × 100/number of spots at 10 μg/mL. (A) Number of IFN-γ SFC for peptides of 9 amino acids within the E57–75 sequence of the YF envelope protein; (B) number of IFN-γ SFC for the 9mers within the E329–343 sequence of the YF envelope protein; (C) IFN-γ ELISPOT evaluation of the affinity of CD8 responses to decreasing in vitro concentrations of 9mers within the E57–75 sequence of the YF envelope protein; (D) IFN-γ ELISPOT evaluation of the affinity of CD8 responses to decreasing in vitro concentration of 9mers within the E329–343 sequence of the YF envelope protein. Panels A and B represent the average of three experiments with a pool of two to three mice. Panels C and D illustrate one representative experiment out of three.
In the second set of 9mer peptides, E332–340 showed the strongest CD8+ stimulatory response with the peptide concentration of 10 μg/mL (Fig. 2D). Contrarily to what was expected, peptide E330–338, which consistently induced the highest number of IFN-γ SFC at 10 μg/mL in the ELISPOT assays, showed only an intermediate stimulatory curve. Peptide E335–343 also showed an intermediate stimulatory activity, while the remaining peptides induced only a very low response, and only at 10 μg/mL.
Taken together, the results pointed out that the 9mer E332–340 was significantly less potent than E330–338 to activate a large number of IFN-γ SFC after in vitro stimulation at 10 μg/mL (Fig. 2B), whereas it could activate relatively higher number of IFN-γ SFC at lower peptide concentrations (Fig. 2D). It suggests that, in animals immunized with the 17DD YF vaccine, peptides E332–340 and E330–338 may be stimulating different populations of CD8+ T cells. The CD8+ T cell population activated by E332–340 seems to be smaller in number, but with a higher avidity for its MHC–peptide complex then the CD8+ T cell population stimulated by E330–338. In these conditions, it is possible that two partially overlapping class I epitopes exist. However, we cannot discard the hypothesis that the in vivo processing and availability of these two epitope are different after the immunization with the 17DD YF vaccine, leading to the priming and generation of different numbers of CD8+ T cell clones with distinct MHC–peptide affinities.
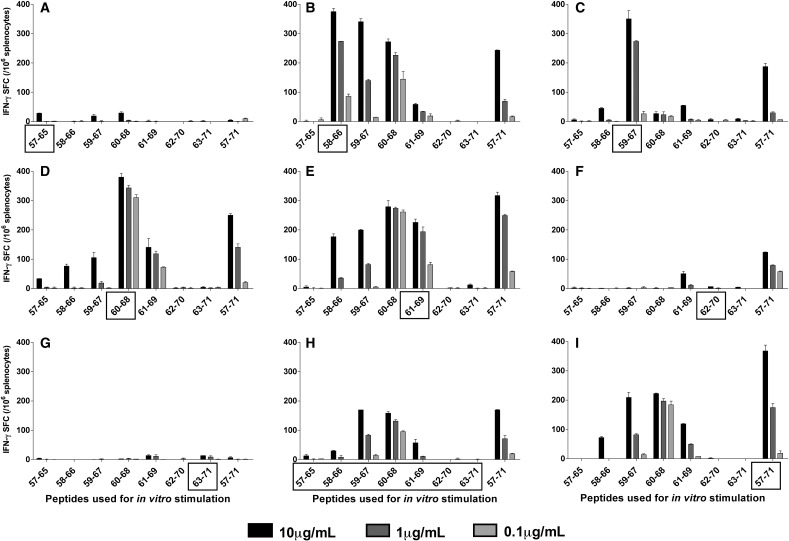
In order to determine whether antigen peptide processing of the 17DD YF virus is affecting the availability of the peptides and therefore influencing the pattern of the in vitro recognition and activation of the cells, mice were immunized with each individual 9mer present in the two sets of immunogenic peptide sequences that produced the highest CD8+ responses, namely E57 to E71 and E329 to E343. Additional control groups were immunized with a mixture of all 9mers pooled together and with the 15mers from which the 9mers were generated. The cells of each group were then tested in ELISPOT assays with decreasing concentrations of all peptides, as means of evaluating the avidity of the CD8 response (Fig. 3 ).
Fig. 3.
Cellular responses to immunization with 9mers from the sequence E57 to E71 of the YF envelope protein. BALB/c mice were immunized s.c. once with 50 μg/mL of individual 9mers, a pool of 9mers, or a 15mer, emulsified v/v in adjuvant (TiterMax®). The splenocytes were used in IFN-γ ELISPOT assays 15–30 days after the immunization. The cells were stimulated in vitro with individual peptides in decreasing concentrations. The box on the x-axis indicates the peptide(s) used in the immunization: Mice were immunized with (A) peptide E57–65; (B) peptide E58–66; (C) peptide E59–67; (D) peptide E60–68; (E) peptide E61–69; (F) peptide E62–70; (G) peptide E63–71; (H) a pool of 9mers; (I) 15mer peptide E57–71. Each plot represents one of three experiments using a single mouse; bars represent the average ± SD of the ELISPOT duplicate.
Among the seven 9mers in the first set, peptides E57–65, E62–70 and E63–71 failed to induce any response after individual immunization (Figs. 3A, F, G). Other four peptides, E58–66, E59–67, E60–68, and E61–69 were able to induce IFN-γ SFC (Fig. 3B–E). Peptide E60–68, as previously observed, showed a very high avidity in the ELISPOT assays, inducing high numbers of IFN-γ SFC at all concentrations of the peptide used for the in vitro stimulation (Fig. 3D). The immunization with peptides that were closely related to peptide E60–68 was also able to induce cells that preferably responded to peptide E60–68, suggesting that a sharing of binding motifs may be the mechanism behind the indirect stimulations. As expected, direct immunization with the 15mer E57–71 also induced a strong response to peptide E60–68, further supporting the concept that this sequence contains the class I motif needed for the CD8 response (Fig. 3I).
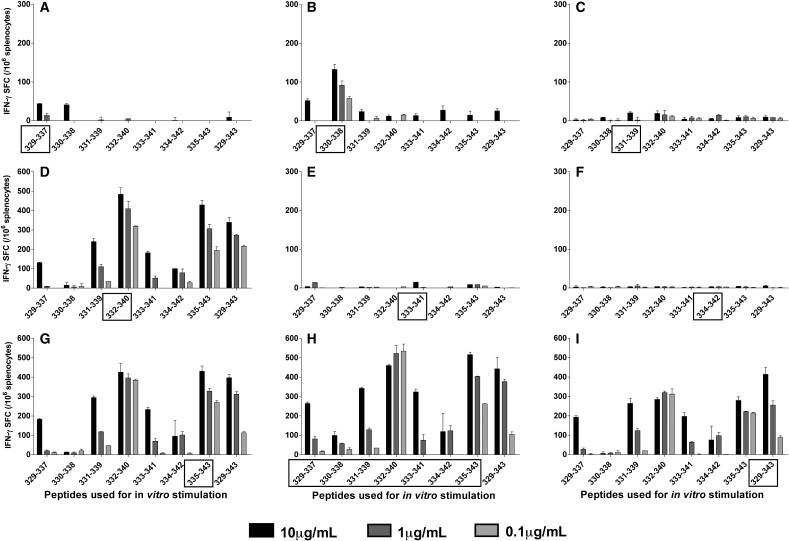
The immunization with the 9mers within the E329 to E343 region produced results distinct of those observed after immunization with 17DD YF virus (Fig. 4 ). The peptides E329–337, E331–339, E333–341, and E334–342 were unable to elicit a significant response, as evidenced by the very low numbers or total lack of IFN-γ SFC after in vitro stimulation with the respective peptides (Figs. 4A, C, E, F). Peptide E330–338, which induced a very strong stimulation after immunization with the 17DD YF vaccine, was less immunogenic when administered as a 9mer peptide emulsified in adjuvant (Fig. 4B). In contrast, the immunization with peptides E332–340 and E335–342 activated relatively higher numbers of IFN-γ SFC (Figs. 4D and G) then the immunization with E330–338, to the in vitro stimulation with the respective peptides. Of note, peptides E332–340 and E335–343 were able to stimulate the secretion of IFN-γ in a cross-reactive fashion, in that both peptides could cross-stimulate the IFN-γ secretion in mice individually immunized with either one of them. Moreover, E332–340 and E335–343 were preferably recognized by splenocytes from mice immunized with the pool of 9mers, as well as those immunized with 15mer E329–343 (Figs. 4H, I). The numbers of IFN-γ SFC obtained with E332–340 were very high, even with the lowest in vitro concentration of 0.1 μg/mL (Figs. 4D, G–I), paralleling the avidity curve previously obtained (Fig. 2D). These results further support the possibility that E330–338 and E332–340 are different epitopes and suggest that either processing of the peptides and/or post-translational modifications of the 17DD YF virus interfere with presentation of these epitopes.
Fig. 4.
Cellular responses to immunization with peptides from E329–343 of the YF envelope protein. BALB/c mice were immunized s.c. once with 50 μg/mL of individual 9mers, a pool of 9mers, or a 15mer, emulsified v/v in adjuvant (TiterMax®). Splenocytes were used in IFN-γ ELISPOT assays 15–30 days after the immunization. The cells were stimulated in vitro with individual peptides in decreasing concentrations. The box on the x-axis indicates the peptide(s) used in the immunization: Mice were immunized with (A) peptide E329–337; (B) peptide E330–338; (C) peptide E331–339; (D) peptide E332–340; (E) peptide E333–341; (F) peptide E334–342; (G) peptide E335–343; (H) a pool of 9mers; (I) 15mer peptide E329–343. Each plot represents one of three experiments using a single mouse; bars represent the average ± SD of the ELISPOT duplicate.
Biochemical characterization of the class I epitopes of the YF envelope protein by binding assay
Subsequently, the affinity between the MHC class I molecules and the immunogenic 9mer peptides were evaluated in binding assays, by directly measuring the affinity of each peptide to all three MHC class I molecules present in the BALB/c mouse strain (Table 1). All eighteen possible 9mers between residues E57 and E75 and between E329 and E340 were assayed to determine their binding affinities to H2-Dd, -Kd, and Ld molecules.
The binding assay results (Table 1) demonstrated that peptide E60–68 has the highest affinity of all the peptides tested, being able to inhibit the binding of the competitor peptide to the H2-Kd molecule at 1 nmol, a result that was consistent with the findings from the ELISPOT assays. In addition, adjoining peptides E59–67 and E61–69, positioned right before and after E60–68 and consequently sharing eight amino acids, also exhibited some degree of affinity for H2-Kd (211 and 93 nmol, respectively); these results could explain why those peptides were also able to stimulate IFN-γ secretion, even without a high avidity, as observed in the ELISPOT assays (Fig. 2C), and also suggest that 8–10mer peptides covering that region can be immunogenic.
Among the 9mers present in the sequence E329 to E343, peptide E332–340 exhibited the highest affinity (IC50 = 56 nmol) for the H2-Ld molecule, while all the other peptides had an IC50 higher than the minimum cutoff value of 500 nmol to be considered a good class I binder (Table 1). Curiously, neither peptide E330–338 nor E335–340 performed well in terms of binding to any of the class I alleles, although they were able to stimulate IFN-γ SFC in the ELISPOT assays with splenocytes of mice immunized with 17DD virus or peptides, even at low peptide concentrations (Fig. 2, Fig. 4).
Characterization of the CD4+ peptides in mice immunized with the 17DD YF vaccine
The seventeen 15mer immunogenic peptides that had been considered positive in ELISPOT assays with splenocytes and with CD8-depleted cells (Fig. 1C) of mice immunized with the 17DD YF vaccine were further characterized. The peptides were tested in ELISPOT assays with decreasing in vitro concentrations (Supplemental Fig. 3). Peptides E57–71, E61–75, E129–143, and E133–147 were able to induce a high CD4+ response even when stimulated in vitro 1 or 0.1 μg/mL; the other peptides showed only an intermediate or low level of stimulatory activity.
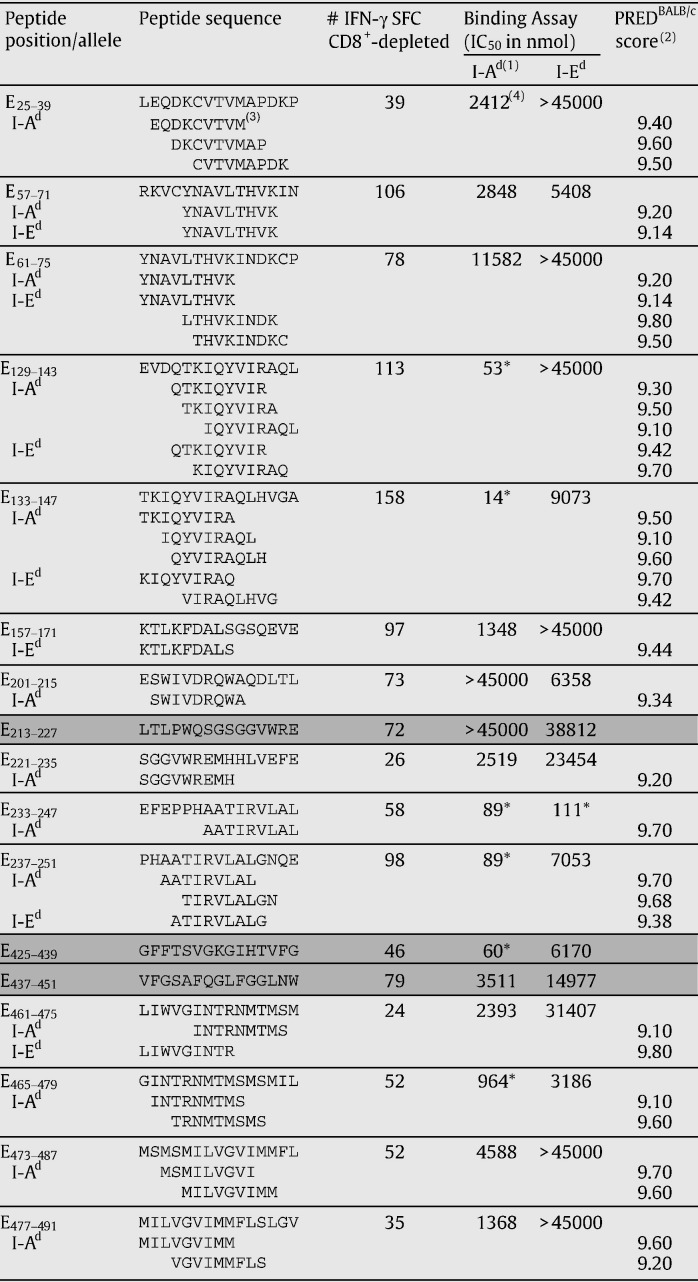
Next, the affinity of the CD4-immunogenic peptides to the H2d class II molecules I-Ad and I-Ed was tested by binding assays (Table 2 ). Peptides E129–143, E133–147, E233–247, E237–251, E325–439, and E465–479 showed affinities lower than 1000 nmol (cutoff for MHC class II binders). Two of them, peptides E129–14 and E133–147, were among the ones that also showed immunogenicity in ELISPOT assays at low peptide concentrations. All other immunogenic peptides tested showed a low affinity in the binding assays (IC50 > 1000 nmol). It is important to note that peptides E129–143 and E133–147, and E233–247 and E237–251 are adjacent and therefore share 11 amino acids, which may explain why both peptides in the two pairs showed similar IC50 values, as well as why they induced high numbers of IFN-γ SFC after the immunization with the 17DD YF vaccine. All six peptides below the cutoff showed a high affinity for I-Ad suggesting that these peptides are being presented by the MHC class II I-Ad molecule. In addition, peptide E233–247, also showed a high affinity for I-Ed (Table 2) suggesting that this peptide may be presented by both, I-Ad and I-Ed molecules.
Table 2.
Results of ELISPOT, binding assay and PREDBALB/c prediction for the CD4-positive ELISPOT 15mer peptides of the yellow fever envelope protein
A list of 94 peptides selected by PREDBALB/c with score above 9 was compared to the CD4 ELISPOT results. The 9mers selected by the prediction software are shown below each 15mer peptide. (1) Molecules of H2d class II; (2) PREDBALB/c software scores; (3) options of 9mer peptides within each 15mers analyzed and scored above 9 by PREDBALB/c; (4) IC50 (nmol) results for the binding assay of H2d class II molecules; ⁎values of IC50 below the cutoff of 1000 nmol, suggestive of a possible binder peptide; Shading — peptides positive for CD4 response by ELISPOT (Fig. 2C) did not predict by PREDBALB/c above a threshold of 9.
Comparison between the ELISPOT results and PREDBALB/c prediction software results for the YF envelope protein
Prediction software is generally used before in vitro testing in order to identify possible MHC binder peptides in a protein or to test the likelihood of a specific peptide inducing an immune response when tested by methods such as ELISPOT. Here, we used PREDBALB/c after our ELISPOT and binding experiments in order to evaluate how well the software would perform in anticipating the positive peptides, as well as in avoiding testing of peptides that did not induce a positive response. Thus, we evaluated the performance of PREDBALB/c to predict possible binders to all the H2d alleles (Dd, Kd, Ld, I-Ad, and I-Ed). We subjected the list of 120 15mer peptides from the envelope protein of the 17DD YF virus (Supplemental Table 2) to PREDBALB/c prediction analysis for class I and class II alleles, independently. It is important to note that in our simulation, the use of either the list of 15mers or the whole sequence of the envelope protein rendered comparable results, possibly due to the high degree of coverage offered by the scheme of 15 amino acids overlapping 11 (Kiecker et al., 2004).
The software analyzed all possible 9mers within each peptide, ranking each peptide according to the highest score found (scores range from 1 to 10) (Zhang et al., 2005). For each one of the alleles, an output result list with 120 scored peptides was generated. Once these lists of peptides were sorted from high to low score, we compared each list to the ELISPOT results (Fig. 1), attempting to visualize the PREDBALB/c score threshold that eliminated the largest number of negative peptides while keeping the positive peptides.
In the case of the class I prediction results for the 120 15mer peptides from the envelope protein, a threshold score of 9 eliminated all positive peptides, while a threshold of 8 included all three positive 15mers observed in the CD8 response (E57–71, E61–75, and E329–434; Fig. 1B). After removing all the repeated peptides in each allele's list, the PREDBALB/c indicated 91 peptides as highly probable binders regardless of the class I allele. If the 91 peptides had been assayed instead of 120 15mer peptides, this approach would have reduced by 24% the number of peptides to test by ELISPOT.
We further evaluated the discrimination of the possible binders found in the PREDBALB/c results by comparing the results produced by the software to all 9mer peptides that were eventually used to characterize the CD8+ response by ELISPOT (Fig. 2), as shown in Table 1. The software correctly predicted E60–68 (score 8.66 for H2-Kd), which was positive by ELISPOT and also presented a high affinity for H2-Kd in binding assays. However, it failed by positively predicting peptide E62–70 (score 8.42 for H2-Dd), which was not positive by ELISPOT or by binding assay. Peptide E331–339 was predicted (score 8.50 for H2-Dd) although it induced only a secondary ELISPOT response and showed no affinity in the binding assay. On the other hand, peptide E330–338, which induced a very strong ELISPOT response, was not predicted by the software (Table 1).
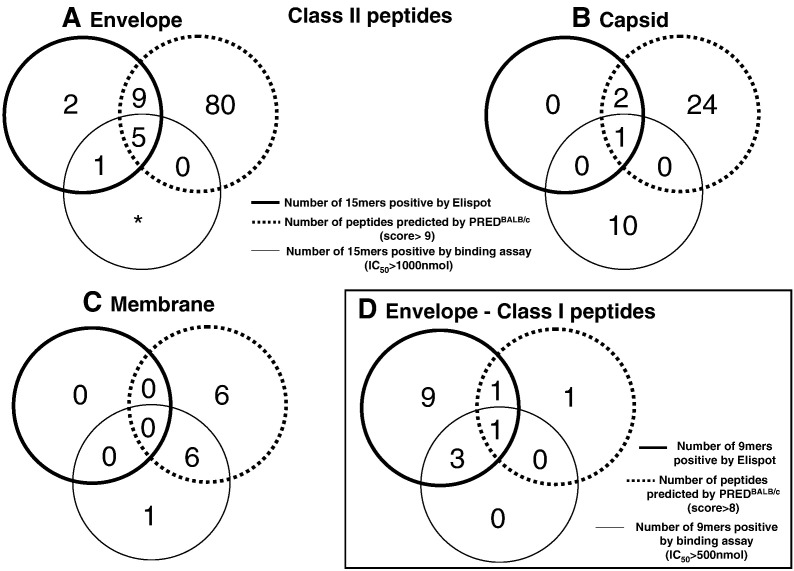
The overall comparison of the ELISPOT results, binding assay results, and PREDBALB/c predictions can be seen in the Fig. 6. The diagram illustrates the differences in immunogenicity as defined by ELISPOT responses, epitope prediction, and binding assays.
Fig. 6.
Venn diagram representation of results obtained from ELISPOT assay binding assay, and PREDBALB/c prediction. (A) ELISPOT results with 120 15mer peptides from the envelope of the YF virus with CD8-depleted splenocytes; PREDBALB/c was used to analyze the 120 15mer peptides from the envelope for the presence of class II (I-Ad and I-Ed) binders; ⁎only the seventeen peptides positive by ELISPOT were analyzed by binding assays for class II molecules, and those showing an IC50 < 1000 nM were considered possible binders. (B) ELISPOT results for the 3 out of 28 15mer peptides from the capsid of the YF virus that were considered positive with CD8-depleted splenocytes; PREDBALB/c was used to analyze the 28 15mer peptides from the capsid for the presence of class II (I-Ad and I-Ed) binders; all 28 15mers from the capsid were tested in binding assays for class II molecules, and those showing an IC50 < 1000 nM were considered possible binders. (C) ELISPOT results with the 16 15mer peptides from the membrane of the YF virus with CD8-depleted splenocytes; PREDBALB/c was used to analyze the 16 15mer peptides from the membrane for the presence of class II (I-Ad and I-Ed) binders; all 16 were tested in binding assays for class II molecules, and those showing an IC50 < 1000 nM were considered possible binders. (D) ELISPOT results for the 18 9mers from the envelope of the YF virus tested with CD4-depleted splenocytes; PREDBALB/c was used to analyze the 18 9mer for the presence of class II (I-Ad and I-Ed) binders; all 9mers were tested in binding assays for class I molecules, and those showing an IC50 < 500 nM were considered possible binders. The intersection represents the number of peptides that were positive in two or three of the approaches.
The analysis of the immunogenic peptide sequences revealed that some of them contained known anchor residues, which in the case of the MHC class I molecules, were consistent with the MHC restrictions found in the binding assays (Supplemental Table 4). However, the MHC class II restriction of 15mer peptides was less evident since anchor motifs for both I-Ad and I-Ed could be found in a same peptide (Supplemental Table 7).
Analysis of the T cell response to the YF virus capsid and membrane protein peptides
As described before, BALB/c mice were immunized twice with the 17DD YF vaccine, 3 weeks apart. The ELISPOT experiments were performed with a set of 15mers, overlapping by 11 amino acids, which comprise the entire length of the capsid (Supplemental Table 3) and membrane proteins (Supplemental Table 4).
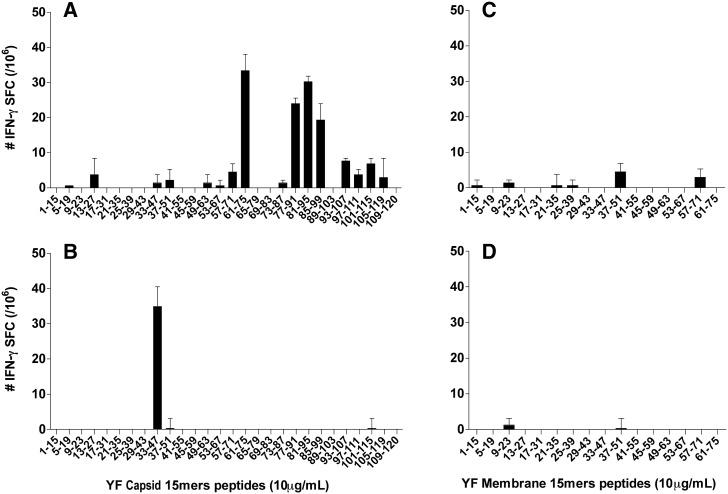
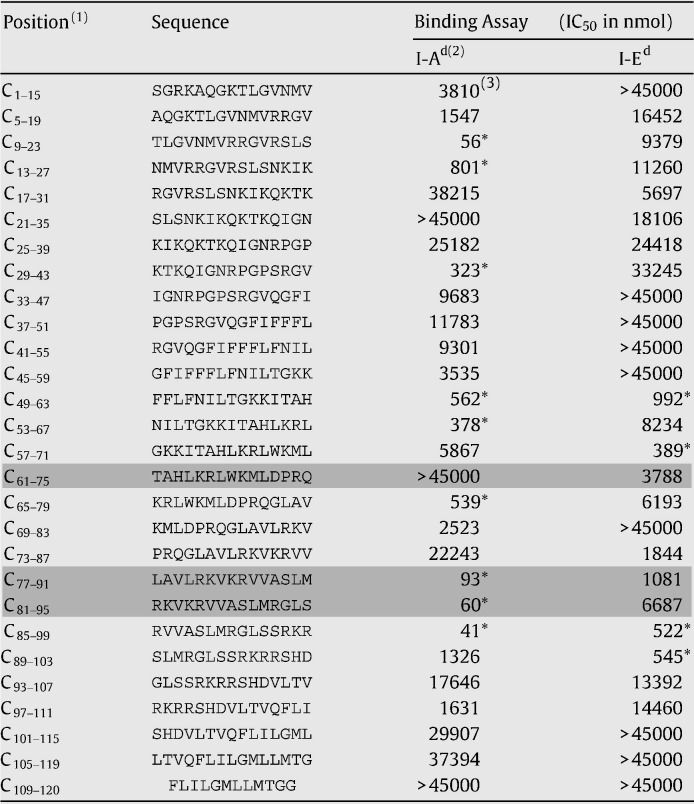
Figs. 5A and B show the profiles of the responses of CD4- and CD8-depleted cells to the capsid peptides in IFN-γ ELISPOT assays. Peptide C33–47 corresponded to the CD8+ response while three other peptides, C61–75, C77–91, and C81–95, were positive for the CD4+ response. Among those, C77–91 and C81–95 showed a high affinity for the MHC class II I-Ad molecule in binding assays (Table 3 ). In addition, other peptides showed a high affinity for MHC class II I-Ad and I-Ed alleles, as seen on Table 3. Unfortunately, the binding properties of the 9mers within the CD8 positive region C33 to C47 were not evaluated because these peptides were not available.
Fig. 5.
Cellular responses of BALB/c mice to the peptides of the YF capsid and membrane protein. BALB/c mice were immunized s.c. with the human 17DD YF vaccine and assessed by ELISPOT assays for the (A) number of IFN-γ SFC for CD8-depleted splenocytes and (B) number of IFN-γ SFC for CD4-depleted splenocytes, using 15mer peptides, overlapping by 11 amino acids from the YF capsid protein (10 μg/mL). (C) number of IFN-γ SFC in CD8-depleted splenocytes and (D) number of IFN-γ SFC in CD4-depleted splenocytes, using 15mers overlapping by 11 amino acids from the YF membrane protein (10 μg/mL). Results represent the mean ± SD from three experiments.
Table 3.
Results of class II binding assay for the 15mer peptides from the yellow fever capsid protein
(1) Position of each 15mer peptide in the capsid protein (C) of the yellow fever virus; (2) molecules of H2d class II (3) IC50 result for the binding assay for H2d class II molecules (nmol); ⁎values of IC50 below the cutoff of 1000 nmol, suggestive of a possible binder peptide; Shading — peptides positive for CD4 response by ELISPOT (Fig. 6A).
The analysis of the capsid 15mer peptides using the PREDBALB/c software yielded 18 peptides above the PREDBALB/c software threshold of 8 (18/28) with probable class I binding motifs and 27 peptides above the threshold of 9 (27/28) with probable class II binding motifs (data not shown). Although the prediction algorithm produced a high number of false-positives for the capsid protein, all the peptides that were defined as immunogenic by ELISPOT were found in the software prediction list. (Fig. 6B).
The number of IFN-γ SFC for the 15mers from the YF virus membrane protein were overall relatively very low in the BALB/c mice immunized with the 17DD YF vaccine as compared to the responses elicited against the envelope protein, and no peptide was considered positive by ELISPOT (Figs. 5C and D). However, the binding analysis of the membrane 15mer peptides for class II molecules indicated that several peptides bound to I-Ad (results on Table 4 ). The PREDBALB/c software predicted 10 peptides for class I epitopes above a threshold of 8 (10/16) and 12 peptides for class II epitopes with a threshold above 9 (data not shown). Among the 12 peptides predicted to have class II epitopes, 6 showed high affinity in the binding assays (Table 4; Fig. 6C).
Table 4.
Results of class II binding assay for the 15mer peptides from the yellow fever membrane protein
| Position(1) | Sequence | Binding Assay (IC50 in nmol) |
|
|---|---|---|---|
| I-Ad(2) | I-Ed(2) | ||
| M1–15 | AIDLPTHENHGLKTR | > 45,000 | 36,639 |
| M5–19 | PTHENHGLKTRQEKW | 13,183 | > 45,000 |
| M9–23 | NHGLKTRQEKWMTGR | 40,852 | > 45,000 |
| M13–27 | KTRQEKWMTGRMGER | 1903 | > 45,000 |
| M17–31 | EKWMTGRMGERQLQK | 20,419 | 20,683 |
| M21–35 | TGRMGERQLQKIERW | 42,531 | > 45,000 |
| M25–39 | GERQLQKIERWFVRN | > 45,000 | 13,242 |
| M29–43 | LQKIERWFVRNPFFA | 593⁎ | 1559 |
| M33–47 | ERWFVRNPFFAVTAL | 7.3⁎ | 2278 |
| M37–51 | VRNPFFAVTALTIAY | 40⁎ | 1917 |
| M41–55 | FFAVTALTIAYLVGS | 6213 | > 45,000 |
| M45–59 | TALTIAYLVGSNMTQ | 102⁎ | > 45,000 |
| M49–63 | IAYLVGSNMTQRVVI | 383⁎ | 29,666 |
| M53–67 | VGSNMTQRVVIALLV | 30⁎ | 32,305 |
| M57–71 | MTQRVVIALLVLAVG | 6484 | > 45,000 |
| M61–75 | VVIALLVLAVGPAYS | 380⁎ | > 45,000 |
(1) Position of each 15mer peptide in the membrane protein of the yellow fever virus; (2) molecules of H2d class II IC50 result for the binding assay for H2d class II molecules (nmol); ⁎values of IC50 below the cutoff of 1000 nmol, suggestive of a possible binder peptide.
Discussion
Others studies have previously analyzed the immune response to the YF vaccine using different immunization protocols with higher doses of the YF virus (van der Most et al., 2002). In those investigations, only a limited number of epitopes were reported; thus, it appears that the immunization protocol or infection model may be an important variable determining the epitope repertoire, and protocol variations can lead to differences in the results obtained. However, it is also possible that differences in the sensitivity of the readout assays used (e.g. flow cytometry versus ELISPOT) as well as the strain of the virus and mouse strain used, could account for the observed results.
The immunogenic CD8 peptides of the YF envelope protein elicited higher numbers of IFN-γ SFC than the immunogenic CD4 peptides did. This pattern was also observed in the memory experiments, in which the peptides inducing responses in CD8+ T cell led to the highest numbers of IFN-γ SFC. The two most immunodominant peptides were able to stimulate both CD8+ and CD4+ T cells to secrete IFN-γ (E57–71 and E61–75) in 17DD YF vaccine immunized mice, likely boosting the immune response by allowing a closer level of collaboration between the cytotoxic and helper arms of the immune response.
The sequence E60–68 was shown to promote the highest number of IFN-γ SFC in 17DD YF vaccine immunized mice. However, other related 9mer adjacent peptides were also immunogenic. It has previously been demonstrated that peptides of 8 to 10 amino acids can bind H2b class I alleles (H2-Kb and -Db) (Moutaftsi et al., 2006), and likely this is also true for H2d class I alleles. The ELISPOT assays using decreasing in vitro concentrations of peptides may provide a measure of the avidity between the MHC–peptide complex and the TCR (Hesse et al., 2001). At the same time, this approach can indirectly reflect the affinity of the TCR–peptide interaction (Hesse et al., 2001), which can lead to distinct cell responses, as well as to different degrees of activation (Wilson et al., 2004). Our results showed that 9mer E60–68 not only induced the highest number of IFN-γ SFC but also had the highest dose–response curve, indicating a very high avidity for the MHC–peptide[E60-68]:TCR complex in 17DD YF vaccine immunized mice. Peptide E64–72 showed an intermediate level of avidity, while all the other 9mers had a very low avidity, supporting the conclusion that the results can be dependent on the in vitro peptide concentration used in the ELISPOT assays. However, a biological role for the responses that were observed only at high in vitro concentrations cannot be disregarded. The additional binding data showed that peptide E60–68 indeed bound the class I allele H2-Kd with very high affinity and that the adjoining peptides, E59–67 and E61–69, also had a high affinity for H2-Kd, despite the fact that they showed a low dose–response curve in the ELISPOT assays.
Peptide E330–338, which induced the high number of IFN-γ SFC in mice immunized with the 17DD YF vaccine, showed only an intermediate dose–response curve with decreasing peptide concentrations in the ELISPOT, suggesting that the MHC–peptide[E330-338]:TCR avidity was low. This peptide also failed to induce a strong immune response when used directly in immunization protocols, and its recognition was inhibited when the immunization was performed in the presence of other peptides covering the adjacent amino acid sequences (Fig. 5H). Moreover, E330–338 also had a low affinity in the binding assay. On the other hand, peptide E332–340, which activated about half of the number of IFN-γ SFC induced by E330–338, showed a very high ELISPOT avidity curve in 17DD YF vaccine immunized mice and had very high affinity for H2-Ld, as determined in the binding assays. Furthermore, peptide E332–340 was able to induce a very strong and dominant immune response when used directly in the immunization protocol with adjuvant (Fig. 4D). These results point out the benefits of performing ELISPOT assays with different concentrations of peptides, after a first round of screening. This approach can identify peptides that only induce responses at high concentration, as well as evaluate the peptide avidity.
The identification of a class I epitope in E329–E343 of the YF virus envelope protein is partially supported by previous work of Rothman and colleagues, who mapped the H2d class I epitope 331SPCKIPFEI339 in the envelope protein of the dengue-2 virus, another member of the flavivirus family (Rothman et al., 1996). Although YF and the dengue virus do not share complete amino acid identity in the 331–339 region of the envelope protein, the presence of a class I epitope at the same position in the envelope protein in both viruses suggests a selective evolution of the immune system to target the same region.
Peptide E335–343, which induced a low number of IFN-γ SFC and presented an intermediate ELISPOT avidity curve after immunization with 17DD YF vaccine (Fig. 2D), was able to induce a vigorous immune response when used to immunize mice directly (Fig. 4G). Interestingly, in the direct immunization with 9mer peptides plus adjuvant, E332–340 and E335–343 induced IFN-γ SFC in a cross-reactive fashion (Figs. 4D and G), even though E335–343 apparently did not seem to be a good MHC binder in the in vitro binding assay. Other studies have demonstrated that the binding affinity in biochemical assays does not always correlate with cytotoxic potential and that, for some epitopes, a lower affinity favors a higher level of cytotoxicity by CD8+ T lymphocytes (Ochoa-Garay et al., 1997). In this sense, the identification of low affinity epitopes can be useful in vaccine development.
Recently, two groups described protective MHC class I restricted-epitopes in the envelope of the West Nile flavivirus in the context of H2b alleles (C57Bl/6 mouse), (Brien et al., 2007, Purtha et al., 2007). There is no data supporting the protective role of the cytotoxic response against YF and it would be interesting to test if the MHC class I epitopes described here can confer some degree of protection to the YF infection. A better characterization of the cellular response against the YF could lead to new vaccine formulations and new parameters of protection besides the humoral response used today.
It is possible that the broader repertoire of the CD4-positive peptides observed here is partially responsible for the strong humoral response observed against the YF envelope protein (Monath, 1986, Monath and Barrett, 2003), which can provide help to B cells that recognize neutralizing epitopes. In fact, Zhao and colleagues have demonstrated that class II epitopes of the SARS coronavirus nucleoprotein are able to support the production of neutralizing antibodies against the virus (Zhao et al., 2007). It is possible that other cytokines such as IL-4 and IL-5 are also secreted in response to other peptides of the envelope protein. Furthermore, the CD4+ T cells activated by peptides of the envelope protein can help support CD8+ T cell cytotoxic clones that are important to the defence against the virus as well as contributing to the memory response (Supplemental Fig. 2) (Johansen et al., 2004, Sun et al., 2004).
The fact that each of the CD4-immunogenic peptides produced different dose–response curves in the ELISPOT experiments suggests that the MHC–epitope complex can exhibit distinct avidities for the CD4+ T cells TCRs (Hesse et al., 2001), a situation that can have important implications in the choice of epitopes for vaccine development. It is important to note that the precise definition of class II epitopes is somewhat less clear than class I epitopes. In fact, mouse MHC class II alleles have been reported to be able to accommodate a variety of motifs (Kurata and Berzofsky, 1990).
It has been previously reported that the envelope protein and non-structural protein 3 (NS3) are the main targets for the immune response against the YF virus (van der Most et al., 2002). Here we have extended these observations by evaluating the cellular response against the three structural proteins of the virus. In fact, the number of immunogenic peptides in the YF envelope protein found in 17DD YF virus inoculated mice was by far larger than the numbers found in the capsid and membrane proteins. These results also underscore the secondary nature of the immune response against the capsid and membrane proteins of the YF virus when compared to the envelope protein.
PREDBALB/c is an in silico peptide identification tool that uses quantitative matrices based on binder and non-binder peptide training datasets to compose equations based on the frequency of amino acids at specific positions within the training set of 9mer peptides (Zhang et al., 2005). In our hands, the software showed to be very sensitive, predicting almost all positive peptides. However, the ability of the software to eliminate negative peptides was low and, if used before the ELISPOT assays, it would have avoided the testing of only about one-quarter of the 120 15mers of the YF envelope protein. In another system, given a list of 41 peptides from the nucleoprotein of the SARS coronavirus, PREDBALB/c predicted 17 peptides with possible class II epitopes, and two peptides were found to be positive by in vitro epitope mapping (Zhao et al., 2007). Taken together, these results suggest that the PREDBALB/c prediction algorithm is indeed very sensitive, but its specificity still needs to be improved.
In conclusion, the results of our epitope mapping of the three structural proteins of the YF virus in BALB/c (H2d) mice suggest that initial computational screening with high thresholds, followed by peptide screening of ELISPOT responses in immunized hosts and by binding assays, may prove to be the most efficient strategy for epitope discovery and analysis. In addition, the results presented here have the potential to identify novel correlates for YF protection in a mouse strain that is commonly used for the validation of YF vaccines.
Materials and methods
YF vaccine and peptides
The human YF vaccine, composed of attenuated 17DD YF virus, was a generous gift from Dr. Ricardo Galler (Oswaldo Cruz Foundation, Rio de Janeiro, Brazil). The vaccine was reconstituted with chilled PBS, kept in an ice bath, and used for immunization of mice within 4 h of reconstitution.
A set of 81 peptides of 16 amino acids (16mers), overlapping by 10 amino acids (16 × 10) and comprising the entire length of the envelope protein (E) of the YF virus (sequence NCBI entry U17066), was synthesized by Synpep (California, USA) (Supplemental Table 1). The purity of this peptide set was stated by the manufacturer to be 70% or lower, it was not HPLC-purified, and the amount of peptide per vial was not provided.
Additional sets of peptides of 15 amino acids (15mers), overlapping by 11 amino acids (15 × 11) and comprising the entire length of the envelope (n = 120) (Supplemental Table 2), capsid (C; n = 28) (Supplemental Table 3), or membrane (M; n = 16) (Supplemental Table 4) proteins of the 17DD YF virus (sequence NCBI entry U17066), were synthesized by Schafer-N (Copenhagen, Denmark). Peptides of nine amino acids (9mers), overlapping by 8 amino acids, were synthesized for regions E57 to E75 (E57–75) and E329 to E343 (E329–343) of the YF envelope protein (Supplemental Table 5). These peptides were HPLC-purified to 80% purity or greater, with the exception of a few peptides that could not be purified and were used as crude products. The identity of each peptide was confirmed by mass spectrometry, and the amount of purified peptide was precisely measured. Stock solutions of all peptides were prepared by dilution in water when possible, or in a solution of 10 to 100% of DMSO, to a final concentration of 20 mg/mL, and stored at − 20 °C. For the ELISPOT assays, the peptides were used at 10, 1, or 0.1 μg/mL as indicated. The maximum DMSO concentration in the ELISPOT assay was 0.05%.
Animals and immunization protocols
Female BALB/c (H2d) mice, 6 to 8 weeks old (Charles River, Kingston, NY), were used in the immunization protocols. They were housed in micro-isolator cages under specific pathogen-free conditions and manipulated according to Johns Hopkins Institutional Animal Care and Use Committee (IACUC) protocol number MO05M336 Three different immunization protocols were used: 1) Mice were immunized three times subcutaneously (s.c.) at the base of the tail with 105 or 104 pfu of the human YF vaccine (strain 17DD, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil) in 50 μL on days 0, 14 and 28. 2) Mice were immunized s.c. with 104 pfu of the vaccine on days 0 and 21. Memory experiments were performed with animals immunized s.c. with 104 pfu of the vaccine on day 0, with or without a boost on day 21, and boosted or not after 4.5 and 9 months. 3) For the evaluation of epitope avidity/specificity by ELISPOT assay, BALB/c mice were immunized once at the base of the tail with 100 μL of an emulsion v/v of TiterMax® adjuvant (CytRx Corporation, Norcross, GA) containing 50 μg of an individual 9mer, a pool of 9mers (50 μg of each), or the 15mer E57–71, E63–75 or E329–343 (50 μg) from the YF envelope protein, or PBS alone. Mice immunized with PBS emulsified in TiterMax® served as a negative control. The experiments were performed 15 and 30 days after the immunization.
ELISPOT assays for enumeration of IFN-γ spot-forming cells (SFC)
Seven to 10 days after the last immunization, the mice were killed and their spleens were removed. Splenocytes were isolated by standard methods, and single-cell suspensions, depleted of red blood cells, were prepared from freshly isolated splenocytes in culture medium (RPMI 1640 medium supplemented with 10% v/v fetal bovine serum, 100 U/mL penicillin/streptomycin, 2 mM l-glutamine, 50 μM 2-mercaptoethanol and 1 M HEPES buffer). Experiments for evaluation of CD4 and CD8 responses were performed with CD8- or CD4-depleted cells, following the procedures from Miltenyi Microbeads kits (Miltenyi, Auburn, California, USA), using LD columns. IFN-γ ELISPOT assays were performed by using an ELISPOT set from BD-Biosciences Pharmingen (San Diego, CA), according to the manufacturer's protocol. Initially, the ELISPOT plates were coated with anti-IFN-γ at 5 μg/mL and incubated at 4 °C/overnight. The plates were blocked with RPMI 1640 containing 10% FCS for 2 h at room temperature, and either total splenocytes (1 × 106 cells/well) or CD8- or CD4-depleted cells (0.5–1.0 ×106 cells/well) from immunized mice were then added. The cells were cultured at 37 °C in 5% CO2 with /mL penicillin/streptomycin, 2 mM l-glutamine), or in the presence of concanavalin A (2.5 μg/mL; Sigma), 109 pfu/mL of inactivated YF virus as a positive control (strain 17DD), or individual 15mers or 9mers from the envelope, capsid, or membrane proteins of the 17DD YF virus at 10 μg/mL, 1 μg/mL, or 0.1 μg/mL, according to the experiment. After 16 h of culture, the plates were washed and incubated with biotinylated anti-IFN-γ for 2 h at room temperature, followed by HRP-conjugated avidin for 1 h at room temperature. Reactions were developed with AEC substrate (Calbiochem-Novabiochem Corporation, San Diego, CA). Final enumeration of IFN-γ SFC was performed using the Immunospot Series 3B Analyzer ELISPOT reader (Cellular Technologies Ltd, Shaker Heights, OH) with aid of the Immunospot software version 3.0 (Cellular Technologies Ltd). The data indicate the number of spot-forming cells (SFC)/106 cells. The results were considered positive if the number of SFC was above 20 and higher than the background (culture with medium alone) plus three standard deviations. The results are presented after subtraction of the background, which was consistently found to be 10 to 35 spots/106 cells throughout the experiments.
Binding assays
Binding of peptides to H-2 allomorphs was determined using quantitative assays based on the inhibition of binding of a radiolabeled standard peptide to purified MHC molecules, essentially as described previously (Sidney et al., 1998). Briefly, 0.1 to 1 nM of a known MHC-binding radiolabeled peptide was co-incubated at room temperature with varying amounts of unlabeled test peptides, 1 µM to 1nM H-2 molecules purified by affinity chromatography, a cocktail of protease inhibitors and, for class I assays, 1 µM human ß2-microglubulin (Scripps Laboratories, San Diego, CA). After 2-day incubation, MHC–peptide complexes were captured on Lumitrac 600 microplates (Greiner Bio-one, Longwood, FL) coated with monoclonal antibody 28-14-8S, SF1-1.1.1, 34-5-8S, MKD6 or 14.4.4 for Ld, Kd, Dd, I-Ad and I-Ed, respectively. Bound radioactivity was measured using a TopCount microscintillation counter (Packard Instrument Co., Meriden, CT). The concentration of peptide yielding 50% inhibition of the binding of the radiolabeled probe peptide (IC50) was then calculated. Peptides were typically tested at six different concentrations covering a 100,000-fold dose range, and in three or more independent assays. Under the conditions utilized, where [label] < [MHC] and IC50 ≥ [MHC], the measured IC50 values are reasonable approximations of the KD values. The peptides used for binding assays were typically ≥ 80% pure (Supplemental Tables 2, 3, 4, and 5).
PREDBALB/c epitope prediction software
PREDBALB/c is a freely available, website-based in silico epitope prediction program (http://antigen.i2r.a-star.edu.sg/predBalbc/) that scans protein and peptide sequences to look for epitopes that are capable of being presented in the context of H2d alleles of the BALB/c mouse strain. It utilizes quantitative matrices that have been validated using experimentally determined binders and non-binders and also by in vivo studies using viral proteins to score and rank proteic sequences of nine amino acids according to their predicted peptide binding to BALB/c major histocompatibility complex (H2d) class I (H2-Kd, -Dd and -Ld) and class II (I-Ed and I-Ad) molecules (41). This software analyzes the sequence of each peptide and scores each one according to the known binding motifs identified for each allele throughout a core sequence of nine amino acids within the peptide sequence. The software supplies a list of all the peptides, ranked from 10 to 0, for each one of the five alleles of H2d. An additional software feature allows the use of a threshold, which can be defined by the user.
For the identification of possible epitopes, PREDBALB/c was given the list of all 15mer peptides used in the ELISPOT experiments: 120 peptides from the envelope, 28 peptides from the capsid, and 16 peptides from the membrane protein, according to the published sequence of the YF virus 17DD (NCBI entry U17066).
Acknowledgments
The authors are in debt with Betty Hart and Delores Henson for the excellent technical support. We thank Dr. Deborah McClellan and Mrs. Claudia Costabile for the editorial review of the manuscript.
Funding: This project has been funded in part with the Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, USA, under the contract N01 AI 40085.
Competing interests: The authors have declared that no competing interests exist.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.virol.2008.04.043.
Appendix A. Supplementary data
Time course and dose–response evaluation of BALB/c mice immunized with human 17DD YF vaccine in terms of the number of IFN-γ SFC obtained after stimulation with YF envelope peptides. BALB/c mice were immunized s.c. with 105 pfu of the human YF vaccine on day 0, 14, and 28, and splenocytes from the mice were tested in IFN-γ ELISPOT assays 2 weeks after each immunization: days 14 (A), 28 (C), and 42 (E), respectively. Other groups of mice were immunized s.c. with 104 pfu of the YF vaccine on days 0, 14, and 28, and splenocytes were again tested in IFN-γ ELISPOT assays 2 weeks after each immunization: days 14 (B), 28 (D), and 42 (F), respectively. The peptides used in the ELISPOT assays were 16mers, overlapping by 10 amino acids and comprising the entire length of the YF envelope protein. Each plot represents the average of three experiments with a pool of three to five mice each; bars indicate the mean ± SD.
T cell memory response to peptides from the YF envelope peptides. Splenocytes from BALB/c mice immunized with the human 17DD YF vaccine were tested in IFN-γ ELISPOT assays using in vitro stimulation with 15mers, overlapping by 11 amino acids and comprising the entire envelope protein. Immunized mice were assayed after 4.5 months (A) or 9 months (B). Immunized mice received a boost after 21 days and were assayed after 4.5 months (C) or 9 months (D). Immunized mice received two boosts, at 21 days and 9 months, and were assayed 7–10 days later (E) or received a boost after 9 months and were tested 7–10 days later (F).
Avidity of the CD4 response to YF envelope peptides as measured by IFN-γ ELISPOT assays. BALB/c mice were immunized s.c. with the human 17DD YF vaccine, and CD8-depleted splenocytes were assayed by ELISPOT for the number of IFN-γ SFC, using decreasing concentrations of 15mer peptides from the envelope protein.
References
- Anthony D.D., Lehmann P.V. T-cell epitope mapping using the ELISPOT approach. Methods. 2003;29:260–269. doi: 10.1016/s1046-2023(02)00348-1. [DOI] [PubMed] [Google Scholar]
- Assarsson E., Sidney J., Oseroff C., Pasquetto V., Bui H.H., Frahm N., et al. A quantitative analysis of the variables affecting the repertoire of T cell specificities recognized after vaccinia virus infection. J. Immunol. 2007;178:7890–7901. doi: 10.4049/jimmunol.178.12.7890. [DOI] [PubMed] [Google Scholar]
- Barba-Spaeth G., Longman R.S., Albert M.L., Rice C.M. Live attenuated yellow fever 17D infects human DCs and allows for presentation of endogenous and recombinant T cell epitopes. J. Exp. Med. 2005;202:1179–1184. doi: 10.1084/jem.20051352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga-Neto U.M., Marques E.T., Jr. From functional genomics to functional immunomics: new challenges, old problems, big rewards. PLoS Comput. Biol. 2006;2:e81. doi: 10.1371/journal.pcbi.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brien J.D., Uhrlaub J.L., Nikolich-Zugich J. Protective capacity and epitope specificity of CD8(+) T cells responding to lethal west nile virus infection. Eur. J. Immunol. 2007;37:1855–1863. doi: 10.1002/eji.200737196. [DOI] [PubMed] [Google Scholar]
- Co M.D., Terajima M., Cruz J., Ennis F.A., Rothman A.L. Human cytotoxic T lymphocyte responses to live attenuated 17D yellow fever vaccine: identification of HLA-B35-restricted CTL epitopes on nonstructural proteins NS1, NS2b, NS3, and the structural protein E. Virology. 2002;293:151–163. doi: 10.1006/viro.2001.1255. [DOI] [PubMed] [Google Scholar]
- Disis M.L., Gralow J.R., Bernhard H., Hand S.L., Rubin W.D., Cheever M.A. Peptide-based, but not whole protein, vaccines elicit immunity to HER-2/neu, oncogenic self-protein. J. Immunol. 1996;156:3151–3158. [PubMed] [Google Scholar]
- dos Santos C.N., Post P.R., Carvalho R., Ferreira I.I., Rice C.M., Galler R. Complete nucleotide sequence of yellow fever virus vaccine strains 17DD and 17D-213. Virus Res. 1995;35:35–41. doi: 10.1016/0168-1702(94)00076-o. [DOI] [PubMed] [Google Scholar]
- Figueiredo L.T. Emergent arboviruses in brazil. Rev. Soc. Bras. Med. Trop. 2007;40:224–229. doi: 10.1590/s0037-86822007000200016. [DOI] [PubMed] [Google Scholar]
- Galler R., Freire M.S., Jabor A.V., Mann G.F. The yellow fever 17D vaccine virus: molecular basis of viral attenuation and its use as an expression vector. Braz. J. Med. Biol. Res. 1997;30:157–168. doi: 10.1590/s0100-879x1997000200002. [DOI] [PubMed] [Google Scholar]
- Hesse M.D., Karulin A.Y., Boehm B.O., Lehmann P.V., Tary-Lehmann M. A T cell clone's avidity is a function of its activation state. J. Immunol. 2001;167:1353–1361. doi: 10.4049/jimmunol.167.3.1353. [DOI] [PubMed] [Google Scholar]
- Hoffmeister B., Kiecker F., Tesfa L., Volk H.D., Picker L.J., Kern F. Mapping T cell epitopes by flow cytometry. Methods. 2003;29:270–281. doi: 10.1016/s1046-2023(02)00349-3. [DOI] [PubMed] [Google Scholar]
- Jennings A.D., Whitby J.E., Minor P.D., Barrett A.D. Comparison of the nucleotide and deduced amino acid sequences of the structural protein genes of the yellow fever 17DD vaccine strain from senegal with those of other yellow fever vaccine viruses. Vaccine. 1993;11:679–681. doi: 10.1016/0264-410x(93)90317-q. [DOI] [PubMed] [Google Scholar]
- Johansen P., Stamou P., Tascon R.E., Lowrie D.B., Stockinger B. CD4 T cells guarantee optimal competitive fitness of CD8 memory T cells. Eur. J. Immunol. 2004;34:91–97. doi: 10.1002/eji.200324231. [DOI] [PubMed] [Google Scholar]
- Kiecker F., Streitz M., Ay B., Cherepnev G., Volk H.D., Volkmer-Engert R., et al. Analysis of antigen-specific T-cell responses with synthetic peptides—what kind of peptide for which purpose? Hum. Immunol. 2004;65:523–536. doi: 10.1016/j.humimm.2004.02.017. [DOI] [PubMed] [Google Scholar]
- Kurata A., Berzofsky J.A. Analysis of peptide residues interacting with MHC molecule or T cell receptor. can a peptide bind in more than one way to the same MHC molecule? J. Immunol. 1990;144:4526–4535. [PubMed] [Google Scholar]
- Monath T.P. In: Pathology of the Flavivirus. Schleisinger S., Schleisinger M.J., editors. Plenum Press; New York: 1986. The Togaviridae and Flaviviridae; pp. 375–440. [Google Scholar]
- Monath T.P. Yellow fever: An update. Lancet Infect. Dis. 2001;1:11–20. doi: 10.1016/S1473-3099(01)00016-0. [DOI] [PubMed] [Google Scholar]
- Monath T.P., Barrett A.D. Pathogenesis and pathophysiology of yellow fever. Adv.Virus Res. 2003;60:343–395. doi: 10.1016/s0065-3527(03)60009-6. [DOI] [PubMed] [Google Scholar]
- Moutaftsi M., Peters B., Pasquetto V., Tscharke D.C., Sidney J., Bui H.H., et al. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat. Biotechnol. 2006;24:817–819. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- Ochoa-Garay J., McKinney D.M., Kochounian H.H., McMillan M. The ability of peptides to induce cytotoxic T cells in vitro does not strongly correlate with their affinity for the H-2Ld molecule: implications for vaccine design and immunotherapy. Mol. Immunol. 1997;34:273–281. doi: 10.1016/s0161-5890(97)00019-9. [DOI] [PubMed] [Google Scholar]
- Peters B., Sidney J., Bourne P., Bui H.H., Buus S., Doh G., et al. The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol. 2005;3:e91. doi: 10.1371/journal.pbio.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland J.D., Calisher C.H., Monath T.P., Downs W.G., Murphy K. Persistence of neutralizing antibody 30–35 years after immunization with 17D yellow fever vaccine. Bull. World Health Organ. 1981;59:895–900. [PMC free article] [PubMed] [Google Scholar]
- Post P.R., Santos C.N., Carvalho R., Cruz A.C., Rice C.M., Galler R. Heterogeneity in envelope protein sequence and N-linked glycosylation among yellow fever virus vaccine strains. Virology. 1992;188:160–167. doi: 10.1016/0042-6822(92)90745-b. [DOI] [PubMed] [Google Scholar]
- Purtha W.E., Myers N., Mitaksov V., Sitati E., Connolly J., Fremont D.H., et al. Antigen-specific cytotoxic T lymphocytes protect against lethal west nile virus encephalitis. Eur. J. Immunol. 2007;37:1845–1854. doi: 10.1002/eji.200737192. [DOI] [PubMed] [Google Scholar]
- Querec T., Bennouna S., Alkan S., Laouar Y., Gorden K., Flavell R., et al. Yellow fever vaccine YF-17D activates multiple dendritic cell subsets via TLR2, 7, 8, and 9 to stimulate polyvalent immunity. J. Exp. Med. 2006;203:413–424. doi: 10.1084/jem.20051720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regner M., Lobigs M., Blanden R.V., Milburn P., Mullbacher A. Antiviral cytotoxic T cells cross-reactively recognize disparate peptide determinants from related viruses but ignore more similar self- and foreign determinants. J. Immunol. 2001;166:3820–3828. doi: 10.4049/jimmunol.166.6.3820. [DOI] [PubMed] [Google Scholar]
- Rothman A.L., Kurane I., Ennis F.A. Multiple specificities in the murine CD4+ and CD8+ T-cell response to dengue virus. J. Virol. 1996;70:6540–6546. doi: 10.1128/jvi.70.10.6540-6546.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sercarz E.E., Lehmann P.V., Ametani A., Benichou G., Miller A., Moudgil K. Dominance and crypticity of T cell antigenic determinants. Annu. Rev. Immunol. 1993;11:729–766. doi: 10.1146/annurev.iy.11.040193.003501. [DOI] [PubMed] [Google Scholar]
- Sette A., Fikes J. Epitope-based vaccines: an update on epitope identification, vaccine design and delivery. Curr. Opin. Immunol. 2003;15:461–470. doi: 10.1016/s0952-7915(03)00083-9. [DOI] [PubMed] [Google Scholar]
- Sette A., Fleri W., Peters B., Sathiamurthy M., Bui H.H., Wilson S. A roadmap for the immunomics of category A–C pathogens. Immunity. 2005;22:155–161. doi: 10.1016/j.immuni.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Sidney J., Southwood S., Oseroff C., del Guercio M., Sette A., Grey H.M. Measurement of MHC/peptide interaction by gel filtration. Curr. Protocols Immunol. 1998;18:11–19. doi: 10.1002/0471142735.im1803s31. [DOI] [PubMed] [Google Scholar]
- Sun J.C., Williams M.A., Bevan M.J. CD4+ T cells are required for the maintenance, not programming, of memory CD8+ T cells after acute infection. Nat. Immunol. 2004;5:927–933. doi: 10.1038/ni1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Most R.G., Harrington L.E., Giuggio V., Mahar P.L., Ahmed R. Yellow fever virus 17D envelope and NS3 proteins are major targets of the antiviral T cell response in mice. Virology. 2002;296:117–124. doi: 10.1006/viro.2002.1432. [DOI] [PubMed] [Google Scholar]
- Wilson D.B., Wilson D.H., Schroder K., Pinilla C., Blondelle S., Houghten R.A., et al. Specificity and degeneracy of T cells. Mol. Immunol. 2004;40:1047–1055. doi: 10.1016/j.molimm.2003.11.022. [DOI] [PubMed] [Google Scholar]
- World Health Organization Yellow fever vaccine. WHO position paper. Wkly. Epidemiol. Rec. 2003;78:349–359. [PubMed] [Google Scholar]
- Yewdell J.W. Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses. Immunity. 2006;25:533–543. doi: 10.1016/j.immuni.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Zhang G.L., Srinivasan K.N., Veeramani A., August J.T., Brusic V. PREDBALB/c: a system for the prediction of peptide binding to H2d molecules, a haplotype of the BALB/c mouse. Nucleic Acids Res. 2005;33:W180–W183. doi: 10.1093/nar/gki479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Huang Q., Wang W., Zhang Y., Lv P., Gao X.M. Identification and characterization of dominant helper T-cell epitopes in the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:6079–6088. doi: 10.1128/JVI.02568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Time course and dose–response evaluation of BALB/c mice immunized with human 17DD YF vaccine in terms of the number of IFN-γ SFC obtained after stimulation with YF envelope peptides. BALB/c mice were immunized s.c. with 105 pfu of the human YF vaccine on day 0, 14, and 28, and splenocytes from the mice were tested in IFN-γ ELISPOT assays 2 weeks after each immunization: days 14 (A), 28 (C), and 42 (E), respectively. Other groups of mice were immunized s.c. with 104 pfu of the YF vaccine on days 0, 14, and 28, and splenocytes were again tested in IFN-γ ELISPOT assays 2 weeks after each immunization: days 14 (B), 28 (D), and 42 (F), respectively. The peptides used in the ELISPOT assays were 16mers, overlapping by 10 amino acids and comprising the entire length of the YF envelope protein. Each plot represents the average of three experiments with a pool of three to five mice each; bars indicate the mean ± SD.
T cell memory response to peptides from the YF envelope peptides. Splenocytes from BALB/c mice immunized with the human 17DD YF vaccine were tested in IFN-γ ELISPOT assays using in vitro stimulation with 15mers, overlapping by 11 amino acids and comprising the entire envelope protein. Immunized mice were assayed after 4.5 months (A) or 9 months (B). Immunized mice received a boost after 21 days and were assayed after 4.5 months (C) or 9 months (D). Immunized mice received two boosts, at 21 days and 9 months, and were assayed 7–10 days later (E) or received a boost after 9 months and were tested 7–10 days later (F).
Avidity of the CD4 response to YF envelope peptides as measured by IFN-γ ELISPOT assays. BALB/c mice were immunized s.c. with the human 17DD YF vaccine, and CD8-depleted splenocytes were assayed by ELISPOT for the number of IFN-γ SFC, using decreasing concentrations of 15mer peptides from the envelope protein.