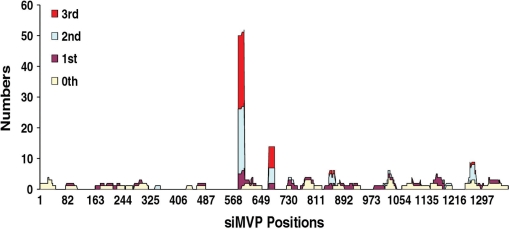
Figure 3.
The positions and numbers of siMVPs identified in each drug selection cycle. HeLa cells expressing the CD-MVP hybrid transcript were incubated with E. coli BT-203 containing the siMVP library for delivery of siMVPs into cells at an m.o.i. of 20 at 37°C for 2 h. After washing as before, 5FC was added to cells at 150 µM concentration. At 48 h after adding 5FC plasmid DNAs containing siMVPs were extracted from surviving cells and transformed into E. coli DH5α. About 30–40 colonies were randomly picked up from the transformants, and plasmid DNAs were reisolated from the cultures of individual colonies for PCR amplification of siMVPs using two primers located the H1 and U6 promoter regions, respectively. The sequences of siMVPs amplified from individual colonies were determined by DNA sequencing. At the meantime, plasmid DNA mixtures were reextracted from the transformed E. coli DH5α, and directly transformed into E. coli BT-203 for the second cycle of bacterial invasion and forward drug selection, and so repeatedly for the third selection cycle.