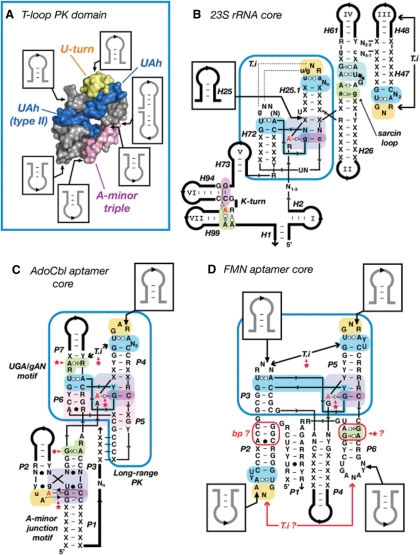
Figure 7.
The T-loop PK domain as a prevalent structural scaffold in natural RNA molecules. (A) Summary of the sequence insertions within the 3D structure of the T-loop PK domain. The constituent sub-motifs are in blue (UA_h), yellow (U-turn) and magenta (A-minor triple). The T-loop PK domain shown is from the 23S rRNA (Hm:301–350; Ec:296–342). (B) Detailed secondary structure diagram of the consensual structural core of the bacterial and Archaea 23S rRNAs deduced from crystallographic structures. Note that the T-loop PK domain (boxed in blue) brings together the various structural domains (I, II, III, IV, V, VI and VII) of the ribosome. (C) Prediction of the architecture of the core of the natural AdoCbl aptamer (62). (D) Prediction of the structural core of the FMN aptamer (62). Positioning of elements P1–P2, P4 and P6 are not well defined compared to P3–P5. The T-loop in P2 should form an intercalating nt interaction with a distant nt that is most likely one of the conserved purines in L6. The GA shared motif in P6 as well as the base pairs in P2 might be involved in one of the riboswitch structural states. (B,C,D) The tertiary interaction (T.i) of the T-loop PK domain (boxed in blue) corresponds to a class of topological equivalence. The conserved T-loop can either recognize another terminal loop (B, D), an UGA/gAN motif (C) or a helix (Figure 2), leading to structurally different but functionally equivalent interactions. Red stars indicate new tertiary interaction predictions [in comparison to (62)]. Thin lines are for zero nt length connectors. Thick lines are for variable length sequence insertions. For annotations, see Figures 2 and S3.