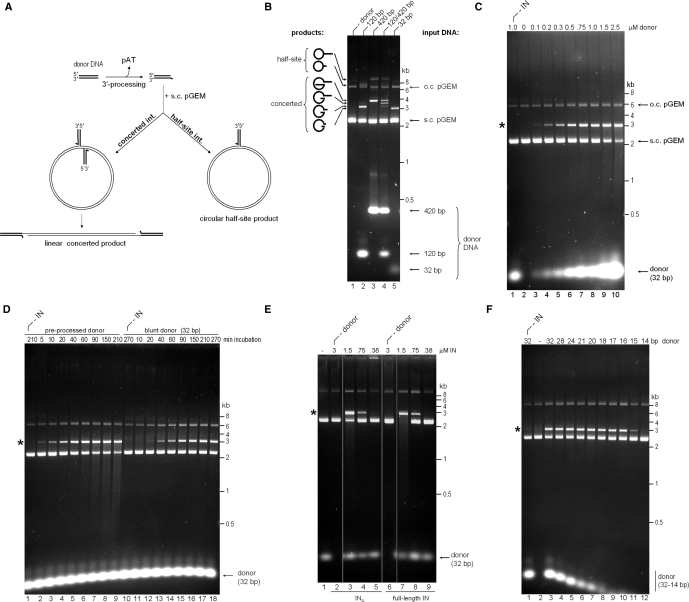
Figure 1.
Strand transfer activity of recombinant PFV IN. (A) Schematic of strand transfer reactions using circular DNA target. Concerted integration results in a gapped linear product, and the half-site process yields tailed relaxed circles. (B) Concerted integration activity of PFV INΔ in vitro. The enzyme (1.5 μM) was incubated with supercoiled pGEM target DNA in the absence (lane 1) or presence of 100 nM donor DNA (lanes 2–5). Blunt DNA fragments of varying lengths mimicking the U5 PFV cDNA terminus served as donors: 120-bp (lane 2), 420-bp (lane 3), an equimolar mixture of 120-bp and 420-bp donors (50 nM each, lane 4), or a 32-bp oligonucleotide (lane 5). Reaction products separated in 1.5% agarose gels were detected with ethidium bromide. Migration positions of concerted and half-site reaction products, open circular (o.c.) and supercoiled (s.c.) pGEM target DNA forms, donor DNA and a DNA size ladder are indicated. (C) PFV INΔ strand transfer activity in the presence of 0–2.5 μM blunt 32-bp donor DNA (lanes 2–10, as indicated). IN was omitted in lane 1. Here and on the remaining panels, the star indicates the migration position of the concerted integration product. (D) Time course of PFV INΔ strand transfer reactions in the presence of preprocessed (lanes 1–9) and blunt (lanes 10–18) 32-bp donor DNA. IN was omitted in lanes 1 and 10. (E) Comparison of strand transfer activities of full-length PFV IN and INΔ. Supercoiled pGEM target DNA was incubated with 0.38–3 μM INΔ (lanes 2–5) or full-length PFV IN (lanes 6–9) in the absence (lanes 2 and 6) or presence (lanes 3–5, 7–9) of preprocessed 32-bp donor substrate; IN was omitted in lane 1. (F) Effect of donor DNA length. Strand transfer reactions using PFV INΔ and preprocessed donors of 32–14 bp, as indicated atop the gel. The enzyme was omitted in lane 1 and donor DNA was omitted in lane 2.