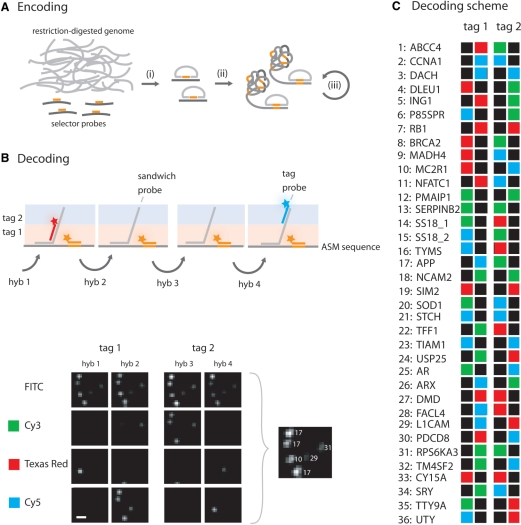
Figure 3.
Multiplex encoding and decoding of genomic loci targeted by selector probes. (A) A genomic sample is prepared by restriction digestion of the sample genome with an appropriate restriction enzyme. (i) Selector probes designed to target specific genomic sequences are added and genomic DNA circles are formed through the action of a thermostable DNA ligase. The circles are either (ii) directly amplified by RCA, or (iii) enriched for by a process which includes restriction digestion of the RCA products into monomers that can be ligated into new circles. The array is created by random immobilization of the ASMs to a poly-l-lysine slide. (B) Arrayed ASMs are decoded by sequential hybridizations of sandwich probes (grey), tag probes (red or blue) and a general tag probe (orange). The sandwich probes contain two regions, one complementary to a specific ASM and one region contains the two decoding tags. These decoding tags, denoted tag 1 and tag 2, are targeted by the tag probes during the hybridization series and their positions in the sandwich probe are illustrated as a pink or blue shade. A small 20 × 20 pixles image subset from one experiment is included, illustrating labeled ASMs after the different hybridization reactions along with an image showing the identified ASMs (scale bar is 2 μm). (C) The decoding scheme used for multiplex decoding of genomic fragments. The names of the gene loci and their corresponding number are listed vertically and the labels from the two tags are illustrated horizontally. Green labels are for Cy3, red labels represent Texas Red and blue correspond to Cy5 labeling. Black means no labeling, i.e. absence of a detectable signal (tag probe).