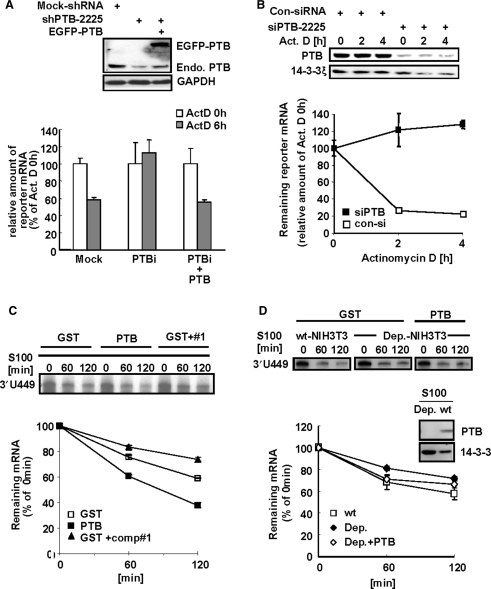
Figure 4.
PTB destabilizes mper2 3′-UTR containing mRNA. (A) HeLa cells were transiently transfected with firefly luciferase reporter construct containing mper2 3′-UTR, β-gal as a normalizing control, and the pShuttle-shPTB plasmid for endogenous PTB knock down, with or without GFP-tagged PTB, and incubated for 24 h before treatment with 5 μM actinomycin D. Total RNA (1 μg) was reverse-transcribed using oligo-dT primer then quantified by real-time PCR. Each value was normalized to β-gal activity. The open bar indicates the mRNA level at time 0 h of actinomycin D treatment and the grey bar indicates the mRNA level 6 h after actinomycin D treatment. The error bars represent the mean ± SEM of three independent experiments. Knocked down and rescued PTB protein levels were validated by western blotting (upper panel). (B) NIH 3T3 cells were transiently expressed with firefly luciferase reporter containing mper2 3′-UTR, β-gal as a normalizing control, and siPTB for endogenous PTB knock down, then incubated for 24 h followed by 5 μM actinomycin D pulse-chase. Total RNA (1 μg) was reverse transcribed using oligo-dT primer then quantified by real-time PCR. Each value was normalized to β-gal activity. Open and closed squares indicate transfection with control siRNA and siPTB-2225, respectively. The error bars represent the mean ± SEM of three independent experiments. Reduction of PTB protein level by siRNA was validated by western blotting (upper panel). (C) In vitro transcribed radiolabeled 3′-UTR probe was incubated with HeLa S100 cytoplasmic extract with or without recombinant PTB (100 nM) or with competitor oligonucleotide #1 (1 μM) for the indicated time. Phenol-chloroform extracted probes were separated in 7 M urea/6%acrylamide gel and subjected to autoradiography (upper panel). Image intensity of each probe was quantified by a phosphoimager. The error bars represent the mean ± SEM of duplicate measurements. (D) In vitro transcribed 3′-UTR probe was incubated for the indicated times with either wild type (wt) NIH-3T3 S100 cytoplasmic extract or PTB-depleted cell extract (Dep.) with or without rescue with recombinant PTB (100 nM). Phenol-chloroform extracted probes were separated in 7 M urea/6%acrylamide gel and subjected to autoradiography (upper panel). Signal intensity was quantified by a phosphoimager (lower panel). The error bars represent the mean ± SEM of duplicate measurements. PTB depletion of the S100 extract was confirmed by immunoblotting (middle panel).