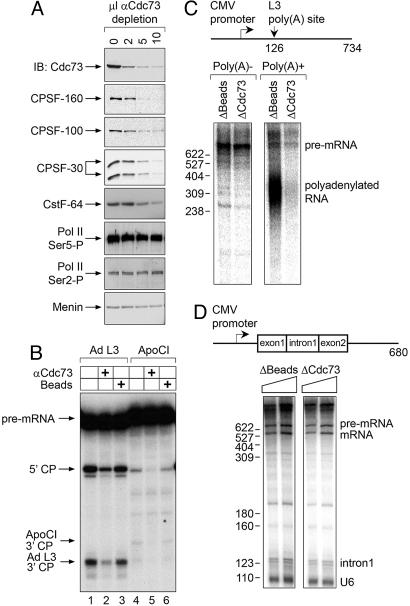
Fig. 2.
The Cdc73 complex is associated with 3′ mRNA processing activity. (A) Nuclear extracts (NE) were immunodepleted with the indicated amounts of anti-Cdc73 Ab648 as shown. Depleted NEs were resolved by gel electrophoresis and immunoblotted with the indicated antibodies. (B) Input NE (lanes 1 and 4), NE after immunodepletion with 10 μL of anti-Cdc73 antibody (lanes 2 and 5), or bead-immunodepleted NE (lanes 3 and 6) were used in an in vitro cleavage assay using labeled adenoviral L3 (Ad L3; lanes 1–3) or human ApoCI (lanes 4–6) substrates. Arrows indicate the precursor mRNA (pre-mRNA), the 5′ cleavage products (5′ CP), and the 3′ cleavage products (ApoCI 3′ CP, and Ad L3 3′ CP). (C) A schematic of the pG3CMVL3 DNA template and the position of the polyadenylation site is shown. Cotranscriptional cleavage and polyadenylation were performed on this DNA template with either bead-depleted or Cdc73-depleted HeLa nuclear extract. RNA products were then separated into nonpolyadenylated (poly(A)−) and polyadenylated (poly(A)+) fractions by oligo(dT) selection, and analyzed on a denaturing gel. (D) At the top is a schematic of the pCMVAdML DNA template. The expected sizes of the pre-mRNA and the processed/spliced mRNA are 680 and 560 bp, respectively. Eight microliters or 10 μL of bead-depleted (Left) or Cdc73-depleted (Right) nuclear extract was used to transcribe and splice the DNA template. RNA products were analyzed on denaturing gel.