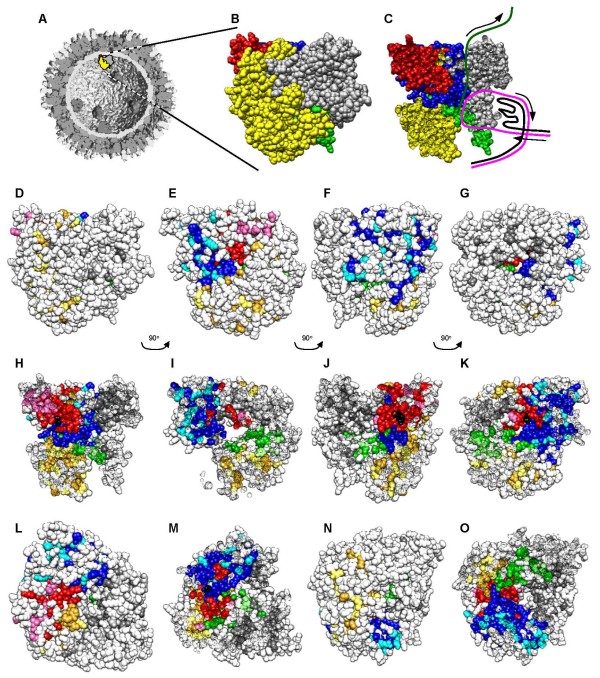
Figure 4.
Localization of conserved, non-conserved, and identical amino acids in ARV, MRV, and AqRV RdRp proteins. The MRV λ3 crystal structure (PDB # 1MUK [52]) was manipulated with Chimera [53]. A, Low-resolution, cutaway model of the reovirus core structure (modified from [26] with permission). B, Blow-up of indicated λ3 molecule in 'A', and C, cut-away of "B" with presumptive paths of genomic (+) RNA (black line), template genomic (-) RNA (magenta line) and nascent mRNA (dark green line) shown (adapted from and as described in [62]); Specific motifs in 'B' – 'O' are color-coded, with N-terminal region in yellow, C-terminal "bracelet" in grey, and canonical polymerase "fingers", "palm", and "thumb" depicted in blue, red, and green, respectively. D, Same as 'B', but in "D" – "O", amino acids that are identical in all 6 ARV, MRV, and AqRV sequences (see Fig. 1) are shown in darker versions of each motif color (goldenrod, dim grey, blue, red, and green, respectively), amino acids that represent conservative substitutions (as determined by Blossum50 matrix) are shown in lighter versions of each motif color (yellow, medium grey, cyan, hotpink, and light green, respectively), and non-conserved amino acids are shown in white. The canonical GDD motif is depicted in black. D - G, represent successive 90° rotations counter-clockwise around vertical axis, of entire RdRp protein, to correspond to front (as depicted in "A"), left side, back, and right side. H - K, represent same views as "D - G", respectively, but with the front approximate half of each view removed. L and N, represent top and bottom view, respectively, of RdRp molecule. M, represents top view, after upper approximately 40% of view removed, and O, represents bottom view, after lower approximately half of view removed. The top surface depicted in "L" is believed to interact with the λ-class core shell protein.