Figure 2.
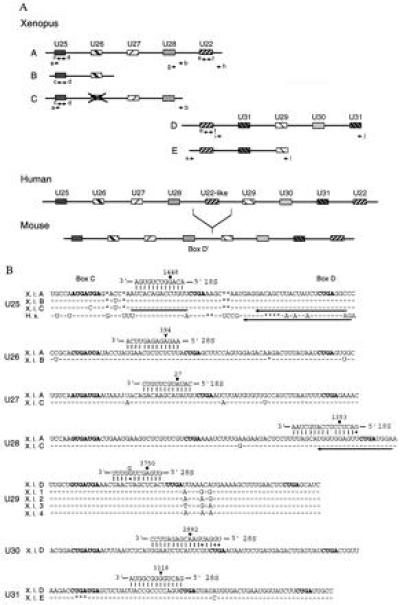
(A) Arrangement of the Xenopus U25–U31 and U22 snoRNA genes (schematic, not drawn to scale). The pairs of arrows indicate primers used for either standard or inverted PCR amplification. Organization of the human and mouse U25–U31 and U22 snoRNA genes (20) are shown for comparison. Boxes filled with identical patterns indicate genes for the same snoRNA. The X over the box in Xenopus fragment C indicates that the U26-like sequence is only 76% identical to human U26 snoRNA and contains several nucleotide changes and deletions within box C and the 18S rRNA complementarity, respectively. (B) Xenopus U25–U31 snoRNA sequences and their predicted base pairing interactions with rRNAs. The sequences of all Xenopus U25–U28, U30, and U31 snoRNA variants, as well as the U29D variant, were deduced from genomic sequences by comparison with their mammalian counterparts, whereas the sequences of the other U29 variants were determined by analyzing their cDNAs. Human U25 is shown for comparison. Identical and missing nucleotides are represented by dashes and asterisks, respectively. Conserved boxes C, D, and D′ are shown in bold. A bar and arrows indicate complementary oligonucleotides used in oocyte injections and for primer extension experiments, respectively. Filled circles or arrowheads indicate rRNA residues that are reported or predicted to be 2′-O-methylated, respectively.
