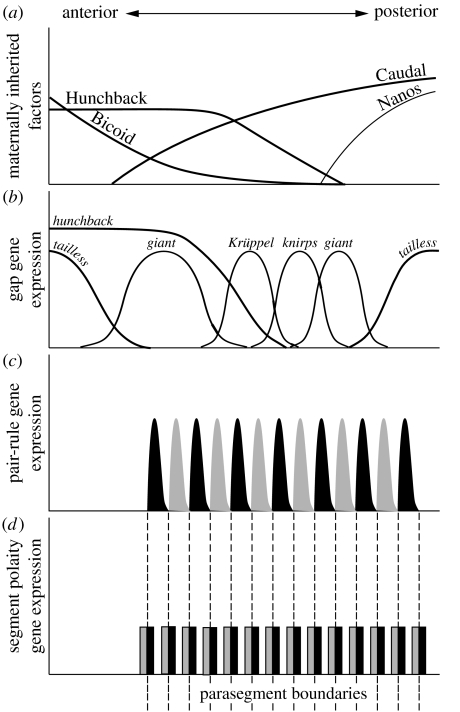
Figure 2.
The Drosophila melanogaster segmentation gene cascade. (a) Maternal genes. Maternal transcripts of the segmentation genes caudal and hunchback are uniformly distributed, whereas maternal bicoid mRNA is tethered to the anterior pole of the egg. Localized at the posterior pole is a complex of maternal proteins and RNAs that includes transcripts of the gene nanos. On fertilization, maternal mRNAs are de-repressed, and Bicoid and Nanos proteins diffuse from their sources of production to form gradients within the egg. Bicoid activates zygotic hunchback expression and represses caudal translation in the anterior, whereas Nanos represses the translation of maternal hunchback in the posterior. As a result Hunchback protein is restricted to the anterior of the egg and protein gradients of Bicoid (decreasing posteriorly) and Caudal (decreasing anteriorly) form. In parallel, a maternally encoded terminal patterning system operates during embryogenesis; the product of torso-like—which is expressed within specialized follicle cells situated at both egg poles during oogenesis—catalyses the localized cleavage of a protein encoded by trunk within the perivitelline fluid. The trunk cleavage product acts as a ligand on the receptor tyrosine kinase encoded by torso, triggering a signalling cascade that regulates the zygotic expression of downstream segmentation genes, such as tailless, at either pole of the egg. (b) Gap genes. The net result of maternal signalling is the activation along the egg antero-posterior axis of a series of zygotic gap genes (i.e. giant, Krüppel, tailless), named so because their mutation leads to gaps in the region of the embryo in which they are normally expressed. The protein products of the gap genes themselves diffuse within the syncytial blastoderm, regulate each other and thus further refine their expression. Gap genes also play an important role at this stage in regulating the expression of the Hox genes, whose protein products confer identity to segments. (c)Pair-rule genes. In the next tier of the Drosophila segmentation cascade are three genes—even-skipped, runt and hairy—whose expression is driven by the maternal and gap gene transcription factor products. All three genes possess complex regulatory sequences that interpret the aperiodic expression of maternal and gap gene products and drive expression in a periodic pattern of seven stripes. These genes are collectively referred to as pair-rule genes since their mutation often leads to abnormalities in alternate segments. The three ‘primary’ pair-rule gene products in turn regulate expression of ‘secondary’ pair-rule genes, such as fushi tarazu, paired, sloppy-paired and odd-skipped. Black curve, even-skipped; grey curve, fushi tarazu. (d) Segment polarity genes. The pair-rule gene products activate the final tier in the Drosophila segmentation gene cascade, the segment polarity genes. These are the genes encoding proteins that actually initiate the formation of segment boundaries, and, as the name suggests, confer polarity to segments. Segment polarity genes are expressed in a series of 14 stripes, with odd and even stripes regulated by a different combination of the pair-rule proteins. The boundary between the expression of two of these genes, engrailed and wingless, becomes the parasegmental boundary, whereas segment boundaries form later, posterior to engrailed expression (adapted from Peel et al. 2005). Black bar, engrailed; grey bar, wingless.