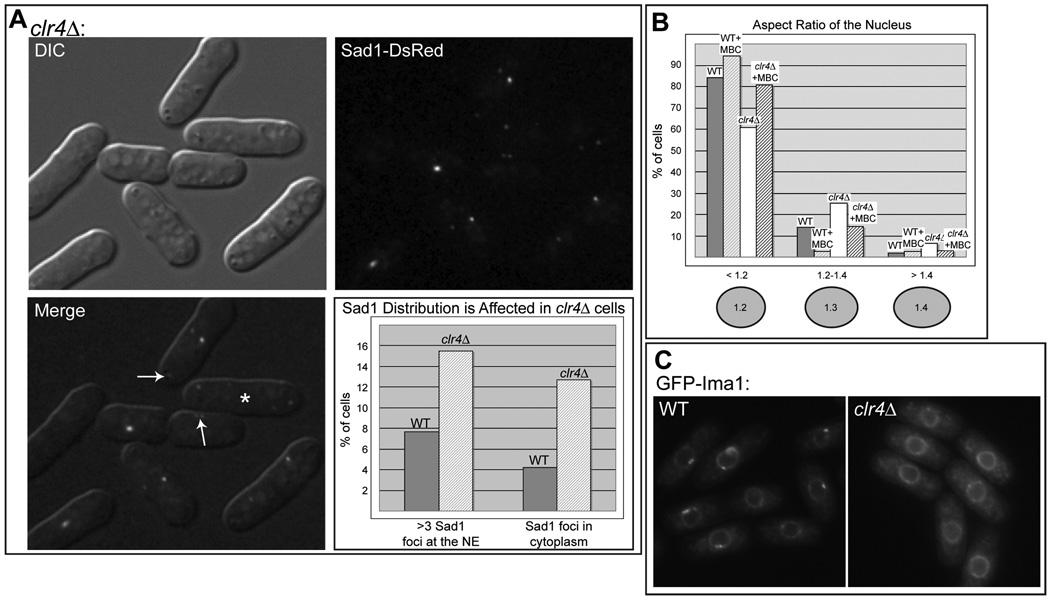
Figure 6. Disruption of histone H3-K9 methylation leads to NE and MAS defects and mislocalization of GFP-Ima1.
A. Sad1 localizes to multiple foci at the NE and to cytoplasmic foci in clr4Δ cells (MKSP123). Fluorescence micrograph showing Sad1-DsRed in the cytoplasm (top arrow). Sad1 is also found fragmented at the NE (lower arrow), including in cells that have recently completed mitosis, which have two Sad1-DsRed foci per nucleus (asterisk). Quantitation of cytoplasmic Sad1 foci was carried out as in Figure 2. B. clr4Δ cells have a mild defect in nuclear shape. Quantitation of nuclear shape was carried out as in Figure 5, using Heh1-GFP as the NE marker (MKSP119). C. The accumulation of Ima1 to the MAS is reduced in clr4Δ cells. Fluorescence micrographs of GFP-Ima1 in WT (MKSP14) and clr4Δ (MKSP120) cells.