Figure 1.
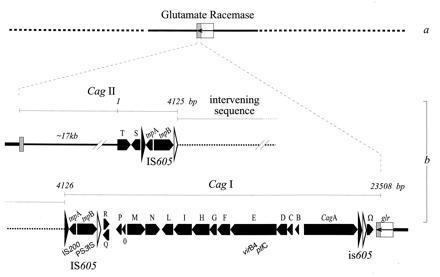
Schematic representation of the cag region as deduced from analysis of strain CCUG 17874. The cag integration site within the glutamate racemase gene (glr) is shaded (a). cag structure: the putative ORFs are represented by arrows (b). Flanking the intervening sequence (dots), the IS605 have a left and right end indicated with large solid and open arrowheads. ORFs with strong similarity are indicated. The cagII region is drawn interrupted but ordered clones and long distance PCR give an approximate indication of the total length, which is indicated by a continuous line. The results of amino acid database searches are included. The name of the genes are in boldface type. LLS, low level of similarity (e−2 > e > e−10); HLS, high level of similarity (>e−50). GenBank, EBI, Swiss-Prot, and Protein Identification Resource releases for December 31, 1995 were used. cagT (LLS), Shigella flexnerii 42-kDa surface antigen IPAC_SHIFL; membrane protein P60 Mycoplasma hominis S42614; Plasmodium falciparum 10b antigen (asparagine rich) J03986 (prokaryotic lipoprotein membrane attachment site). cagS (LLS), Erwinia chrysantemi IIABC component PTS system PTBA_ERWCH; Clostridium perfringens transposase for IS115 TRA1_CLOPE·tnpA (HLS), IS200 from E. coli, S. typhimurium, and Yersinia pestis L25848, U22457, and L25845; S. typhimurium plasmid pSDL2/spvD26 (vsdA–F genes for virulence proteins) X56727. tnpB (HLS), thermophilic bacterium PS3 gene for transposase-like protein D38778; S. typhimurium virulence protein vsdF P24421P24421. cagR (LLS), preprotein translocase secY subunit SECY_STACA; polysialic acid transport protein KPSM_ECOLI; cagQ (LLS) Neisseria gonorrhoeae prepilin peptidase P33566; glutamate/aspartate transporter E. coli GLTJ_ECOLI; 42-kDa membrane antigen precursor Shigella IPAC_SHIFL. cagP (LLS), fimbrial assembly protein (serogroup I) Fmbi_BACNO; type 4 prepilin-like protein-specific leader peptidase LEP3_PSEAE; hydrophobic membrane protein Haemophilus U32720. cagO (LLS), transport system permease P69_Mychr; polysialic acid transporter E. coli KPSN_ECOLI; NADH-ubiquinone oxidoreductase chain 5 (also chain 1, 2) NU1M_ASCSU. cagM (LLS), hook-associated protein type 3 U12817; merozoite surface antigen PFMEZSA1A_1. cagN (LLS), Lmp 1 M. hominis U21963; acidic basic repeat antigen P. falciparum A12521; α-cardiac myosin hc M76598. cagL (LLS), preprotein translocase SECA_ANTSP; proteases secretion protein PRTE_ERWCH. cagI (LLS), CFA/I fimbrial subunit D CFAD_ECOLI; FlaB C. coli A35146;Treponema membrane protein precursor P29721. cagH (LLS), extracellular protease precursor PROA_XANCP; GSP protein d precursor Erwinia chrisantemi GSQD_ERWCH;Treponema membrane protein precursor P29721. cagG (LLS), flagellar motor switch protein FLIM_CAUCR; toxin coregulated pilus biosynthesis protein D (TCP pilus biosynthesis protein TCPD)_VIBCH; GSP protein d precursor Erwinia chrisantemi GSPD_ERWCH. cagF (LLS), ToxB, ToxA_CLODI. cagE (HLS), VirB4 homolog_BPERT (ptlc); VirB4 homolog (plasmid pTiA6)_AGRT9; TraB of IncN plasmid pKM101; TrbE (Tra2 region) of IncP plasmid RP4 (Walker box). cagD (LLS), flagellar hook-associated protein 2 (filament cap) FLID_SALTY; surface presentation SPAO_SHIFL. cagC (LLS), TRAH protein precursor TRH1_ECOLI; SECE_ECOLI protein-export (preprotein translocase); 62-kDa membrane antigen IPAB_SHIDY; sensor protein ENVZ_SALTY. cagB (LLS), sensor protein UHPB_ECOLI; hypothetical 16.6-kDa protein outside the virF region (ORF3)_AGRT9; BACN17G_7 hypothetical protein BACSU; transport system permease protein p69_MYCHR. glr (HLS), glutamate racemase LEPRAE_BREVIS_COLI. Coordinates, headers, and comments are included in the submitted sequence.
