Figure 1.
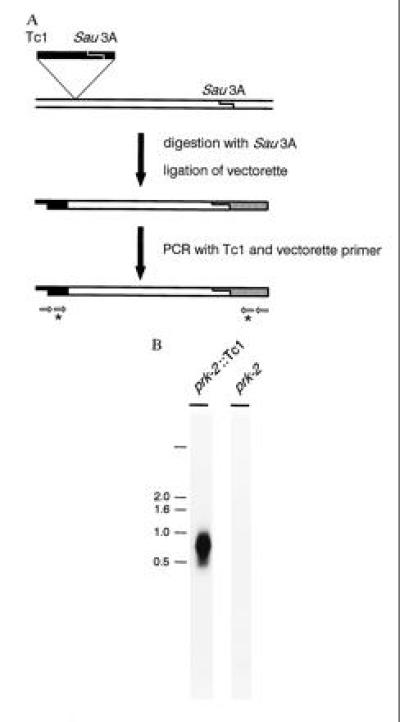
Amplification of Tc1 flanking genomic sequence using an anchored PCR based method. (A) Schematic representation of the anchored PCR based approach. Genomic DNA was digested with Sau3A, resulting in small fragments consisting of Tc1 sequence and flanking genomic sequence. To amplify these fragments, a vectorette oligonucleotide anchor was ligated to the digested DNA. The Tc1 flanking fragments were amplified using primers that anneal to Tc1 and the vectorette anchor. Note that vectorette anchors can ligate at both ends of the fragments. The vectorette is, however, constructed in such a way that the vectorette PCR primers can only anneal after a complementary strand has been generated in the first round of PCR by synthesis from the Tc1-specific primer. Therefore, fragments containing only vectorette anchors, but no Tc1 sequence are not amplified. ∗, Internal Tc1 and vectorette primers containing restriction sites for EcoRI and SphI, respectively. (B) Southern blot analysis of the total vectorette PCR product using genomic DNA of a strain with a Tc1 insertion in the gene prk-2, compared with a similar strain without this insertion. Hybridization with a 32P-labeled prk-2 genomic probe shows specific amplification of a prk-2 fragment in the prk-2::Tc1 strain but not in the strain without this insertion.
