Abstract
Group 1.1c Crenarchaeota are the predominating archaeal group in acidic boreal forest soils. In this study, we show that the detection frequency of 1.1c crenarchaeotal 16S rRNA genes in the rhizospheres of the boreal forest trees increased following colonization by the ectomycorrhizal fungus Paxillus involutus. This effect was very clear in the fine roots of Pinus sylvestris, Picea abies, and Betula pendula, the most common forest trees in Finland. The nonmycorrhizal fine roots had a clearly different composition of archaeal 16S rRNA genes in comparison to the mycorrhizal fine roots. In the phylogenetic analysis, the 1.1c crenarchaeotal 16S rRNA gene sequences obtained from the fine roots formed a well-defined cluster separate from the mycorrhizal ones. Alnus glutinosa differed from the other trees by having high diversity and detection levels of Crenarchaeota both on fine roots and on mycorrhizas as well as by harboring a distinct archaeal flora. The similarity of the archaeal populations in rhizospheres of the different tree species was increased upon colonization by the ectomycorrhizal fungus. A minority of the sequences obtained from the mycorrhizas belonged to Euryarchaeota (order Halobacteriales).
Nonthermophilic Crenarchaeota colonize several types of soils and rhizosphere environments (e.g., references 12, 18, 22, 34, and 40). Several studies report that nonthermophilic Crenarchaeota represent between 0.5% and 3% of the total prokaryotic populations in different soils (4, 21, 30). In most studied soils, the predominant type of crenarchaeotal 16S rRNA gene detected is group 1.1b of the uncultivated nonthermophilic Crenarchaeota, but group 1.1a Crenarchaeota have also been found in some soil environments (5, 7). Nevertheless, group 1.1a Crenarchaeota are more frequently found in marine and other aquatic habitats (6, 14, 25). Group 1.1c Crenarchaeota appear to be restricted to more-specific environments, as they have presently been found mainly in acidic boreal forest soils (2, 3, 10, 12, 40).
Recent studies show that in addition to boreal forest soils, group 1.1c Crenarchaeota also inhabit the soil of the Austrian alps (19, 20), although group 1.1b Crenarchaeota are more abundant. In the alpine soil, only group 1.1b Crenarchaeota were found in young soil uncovered by ice for only 4 years and lacking significant vegetation. However, 1.1c Crenarchaeota formed an integral part of the community in soil that had been free from ice cover for over 135 years and had a stable vegetation cover. An even richer diversity of 1.1c Crenarchaeota was observed in 9,500-year-old vegetated soil. For the vegetated and certainly mycorrhizospheric soils of the Rocky Mountains (Colorado), Oline et al. (22) showed that group 1.1c Crenarchaeota colonized the soil from the foot hill forest (1,800 m above sea level) to the alpine tundra (3,400 m above sea level), but 1.1b Crenarchaeota were still the most abundant archaea.
Crenarchaeota have been found to be associated with the roots of a variety of plant species (5, 8, 31). In a recent study (34), archaea were surveyed in roots of 13 different plant genera, collected from their natural habitats, including turf, prairie, agricultural, and forest sites in a temperate region. All the archaea detected belonged to the 1.1b group, and the diversity of these archaea did not vary significantly between plant genera from the same site, only between plant roots and bulk soil. There was more variability between the different geographical locations than between the plant genera.
Pinus sylvestris is one of the dominant tree species in Finnish forests. When grown in boreal forest humus, nonmycorrhizal P. sylvestris fine roots harbor high numbers of bacteria (37), but they appear to be nearly devoid of archaea (2, 3). In nearly all cases in our previous work, archaea were detected in P. sylvestris fine roots only when they were colonized by mycorrhizal fungi (3). Sliwinski and Goodman (34) were able to show archaeal group 1.1b colonization in roots of Pinus species in temperate forests but did not report the mycorrhizal status of the sampled roots.
In the present study, we surveyed the archaeal population compositions in two coniferous and two deciduous tree species developed in boreal forest humus. As the ectomycorrhizal fungus Paxillus involutus is symbiotic with all four tree species, it was used to test the effect of mycorrhizal colonization on the archaeal populations on the roots.
MATERIALS AND METHODS
Synthesis of test mycorrhizospheres.
Mycorrhizospheres were produced in thin microcosms containing sieved boreal forest humus, as previously described (2). The humus was retrieved from a dry pine forest in southern Finland (60°28′N, 23°45′E). In this experiment, we used four different tree species [Pinus sylvestris L., Picea abies (L.) H. Karst., Betula pendula Roth, and Alnus glutinosa (L.) Gaertner], all of which are common in Finnish forests. P. sylvestris and P. abies are evergreen coniferous trees, and B. pendula and Alnus glutinosa are deciduous trees, which drop their leaves in the autumn. Ten replicate microcosms of each species were prepared, each microcosm containing one individual seedling. Prior to being planted, the tree seedlings were grown aseptically for 1.5 months in 100-ml test tubes containing sterile expanded-clay pellets (Leca) (36). Five seedlings of each tree species were inoculated with the ectomycorrhiza-forming fungus Paxillus involutus Batsch: Fr. (isolate PIL1). A total of 40 microcosms were grown for 1.5 to 2.5 months, with a 20-h photoperiod, a photon influence rate of 250 μmol m−2 s−1, and temperatures of 15 to 20°C for shoots and ca. 10°C for roots. Microcosms were kept moist with a distilled water spray.
Ten individual mycorrhizal root tips (0.02 g [fresh weight {fw}]) were collected from each P. involutus inoculated seedling, and 10 to 15 individual fine roots (0.02 g [fw]) were collected from each noninoculated seedling. The mycorrhizas or fine roots obtained from one seedling were considered one mycorrhizal and one fine-root sample, respectively. Totals of 20 mycorrhizal and 20 fine-root samples, representing five replicates of each tree species, were collected with sterile forceps from the microcosms for DNA extraction. Five samples of 0.2 g (fw) uncolonized forest humus outside the mycorrhizosphere were also harvested from different microcosms. The samples were kept at −80°C until DNA extraction.
DNA extraction and PCR.
The mycorrhizal and fine-root samples were ground with sterilized quartz sand in 50 μl sterile double-distilled water to disrupt the plant and fungal tissues prior to DNA extraction. The humus samples did not require any preextraction treatment. DNA was extracted with a MoBio soil DNA extraction kit (MoBio Laboratories, Inc.) according to the manufacturer's instructions.
A nested PCR was performed with an Eppendorf MasterCycler gradient (Eppendorf) to detect archaeal 16S rRNA genes. To test the effects of primer combinations, PCRs were carried out with three different primer sets on all samples (Table 1). All primer sets targeted sequences at the beginning (forward primers) and middle (reverse primers) of the 16S rRNA gene. All primers used in this study were designed by others to be specific to and target most archaea. PCRs were performed with Phusion proofreading PCR polymerase (Finnzymes) according to the manufacturer's instructions. The sizes and quality levels of the PCR products were checked with 1% agarose-ethidium bromide gels according to standard protocols (29).
TABLE 1.
Primer combinations used in PCR
| Combination no., taxon, and primer name | Primer sequence | Positions (E. coli numbering) | Source or reference | No. of different OTU types (taxon)a
|
||
|---|---|---|---|---|---|---|
| Fr | Mm | Uh | ||||
| 1 | ||||||
| Universal | 8 (Crenarchaeota) | 23 (Crenarchaeota) | ||||
| 8f (fD2) | 5′-AGAGTTTGATCATGGCTCA-3′ | 8-26 | Modified from reference 39 | 7 (Bacteria) | 2 (Bacteria) | |
| 1512r (rP2) | 5′-CCGGCTACCTTGTTACGACTT-3′ | 1492-1512 | 7 (unidentified) | 6 (unidentified) | ||
| Archaea | ||||||
| A109a | 5′-ACKGCTCAGTAACACGT-3′ | 109-125 | 8 | |||
| A934b | 5′-GTGCTCCCCCGCCAATTCCCT-3′ | 915-934 | 35 | |||
| 2 | ||||||
| Archaea | 7 (Crenarchaeota) | 7 (Crenarchaeota) | 3 (unidentified) | |||
| Ar3f | 5′-TTCCGGTTGATCCTGCCGGA-3′ | 7-26 | 12 | 2 (Bacteria) | 2 (Euryarchaeota) | |
| Ar9r | 5′-CCCGCCAATTCCTTTAAGTTTC-3′ | 906-927 | 12 | |||
| Archaea | ||||||
| A109a | 5′-ACKGCTCAGTAACACGT-3′ | 109-125 | 8 | |||
| A934b | 5′-GTGCTCCCCCGCCAATTCCCT-3′ | 915-934 | 35 | |||
| 3 | ||||||
| Archaea | 4 (Crenarchaeota) | |||||
| Ar3f | 5′-TTCCGGTTGATCCTGCCGGA-3′ | 7-26 | 12 | 5 (unidentified) | ||
| A934b | 5′-GTGCTCCCCCGCCAATTCCCT-3′ | 915-934 | 35 | |||
| Archaea | ||||||
| A109a | 5′-ACKGCTCAGTAACACGT-3′ | 109-125 | 8 | |||
| Ar9r | 5′-CCCGCCAATTCCTTTAAGTTTC-3′ | 906-927 | 12 | |||
Fr, fine root; Mm, mycorrhiza; Uh, uncolonized humus.
DGGE analysis.
Nested PCR products of approximately 800 bp were separated by denaturing gradient gel electrophoresis (DGGE) in a 6% acrylamide-bis (37.5:1) gel with a 40 to 60% urea-formamide gradient. The electrophoresis was run at 65 V for 17 h at 60°C. The gels were stained with Sybr green II (Promega) according to the manufacturer's instructions. The gels were imaged with a GelDoc XR UV light imager (Bio-Rad). Bands were divided into groups according to their mobility compared to that for the 1-kb molecular size standard (Invitrogen). Each gel was run with three standard lanes, one in the middle of the gel and one on each side of the gel. The mobility of each DGGE band was compared to that for the standard lanes, and DGGE bands having the same mobility were considered to belong to a group of operational taxonomic units (OTUs). The most prominent bands were excised from the gels with a sterile scalpel, and the DNA was eluted using the “crush and soak” method (23).
Sequencing.
Isolated DGGE fragments were reamplified for sequencing with the same nested primer pair used for the original amplifications (A109a/A934b or A109a/Ar9r) and sequenced at the Haartman Institute, University of Helsinki, with primers Ar514r (11) and Ar344f (28). The sequences obtained with these primers overlapped in the middle of the fragment, as they were produced from the middle toward the ends of the gene fragment (as in reference 22). This method also resulted in sequences of different lengths, as the signal of the sequencing reaction subsided variably. After an initial sequencing of one representative band from each OTU group, two or more comigrating bands from different samples of the same type, obtained with the same primer combination, were chosen for sequencing to test the reproducibility of the DGGE method. The sequences were edited with the program package Vector NTI (InforMax) and compared to all archaeal and bacterial 16S rRNA gene sequences in the DDBJ/EMBL/GenBank databases with BLASTn. Most sequences obtained were 450 to 700 bp. The sequences were tested for chimeras with the Chimera Check tool in Ribosomal Database Project II (http://rdp.cme.msu.edu/index.jsp). No chimerical sequences were found, but all unspecific and bacterial sequences were subtracted from the analysis.
Phylogenetic analyses.
The edited sequences were aligned using the ClustalW alignment tool (http://www.ebi.ac.uk/) with default settings, except that the Gonnet matrix was used. The alignments were checked and manually edited with the BioEdit alignment editor (version 7.0.5; Tom Hall). Maximum-likelihood and maximum-parsimony analyses were performed with PAUP 4.0b (Sinauer Associates, Sunderland, MA), using heuristic search parameters for 10 repeats of stepwise addition of random sequence with a tree bisection-reconnection branch-swapping algorithm. A maximum of 10,000 trees was set for the maximum-parsimony analyses. Consensus bootstrap trees were calculated for both maximum-parsimony and maximum-likelihood phylograms with 1,000 bootstrap replicates. For the initial phylogenetic analyses, all sequences were used. However, some of the sequences were too short to be used for reasonable phylogenetic analyses and were left out of the final analysis.
Statistical analyses.
Statistical significance for differences between frequencies of occurrence of archaeal OTUs in different sample types was determined by Kruskal-Wallis nonparametric one-way analysis of variance and post hoc analysis using Dunn's multiple comparison test (InStat package; GraphPad Software, Inc.). The tests were performed on the combined results for OTUs obtained by all three primer sets. Nonarchaeal and unidentified OTUs were omitted from the analysis.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the new sequences are AM903346 to AM903374.
RESULTS
Detection of archaea by PCR.
PCR products containing archaeal 16S rRNA gene sequences were retrieved from 32 of 45 samples. No PCR products were amplified from P. sylvestris fine-root samples, whereas P. abies, B. pendula, and A. glutinosa fine roots gave positive PCR amplification products (Fig. 1). PCR products of the expected size were retrieved from two of the five uncolonized humus samples and all mycorrhizal samples.
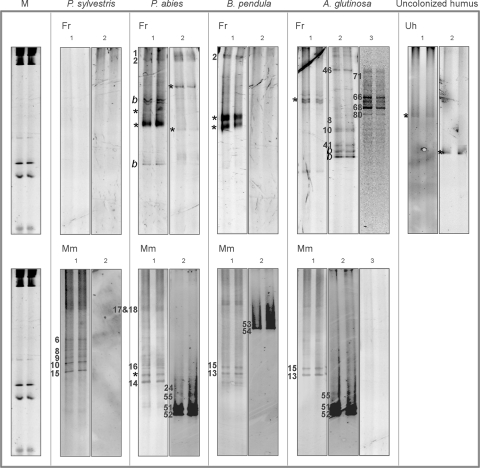
FIG. 1.
Representative lanes of DGGE runs of all different sample types. Samples in the upper sectors are from fine roots (Fr; nonmycorrhizal), and those in the lower sectors are from mature mycorrhizas (Mm; inoculated with P. involutus). The tree species is marked above each sector, with the sector furthest to the right representing uncolonized humus (Uh) samples. A molecular marker (M) is shown, and the primer combination numbers (Table 1) are marked above each pair of lanes. The archaeal bands are marked with a number corresponding to the OTU codes. b, bacteria; *, unidentified.
The different primer combinations showed a clear variation in levels of PCR success as well as in OTU (DGGE band type) richness in the different samples (Table 1 and Fig. 1). Primer combination 1 showed the highest number of different archaeal OTUs in this study. However, the specificity to archaea was lowered by using universal first-round primers. The effect was particularly clear in fine roots, where 7/22 detected DGGE bands were bacterial. Primer combination 2 produced only 2 bacterial OTUs among the 21 obtained OTUs, both from A. glutinosa fine-root samples. Primer combination 3 produced exclusively archaeal OTUs. Only archaeal OTUs were submitted for statistical analysis. Upon sequencing, no redundancy was detected between the OTUs produced with the different primer pairs.
Comparison of average appearances of archaeal OTUs.
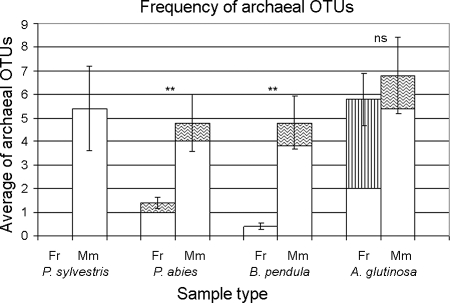
The number of archaeal OTUs was statistically significantly higher in mycorrhizal samples than in fine-root samples of P. abies and B. pendula (two-tailed P = 0.007 and 0.003, respectively) (Fig. 2). The result was the same for P. sylvestris, but statistical analyses were not carried out, due to a complete absence of archaeal OTUs in fine roots. The same trend could be seen in the A. glutinosa samples with primer combinations 1 and 2, although the result was not statistically significant (two-tailed P = 0.21). Primer combination 3 specifically amplified archaeal 16S rRNA gene sequences from A. glutinosa fine-root samples, resulting in a high average number of OTUs compared with the levels for other fine-root samples. Nevertheless, the difference in number of archaeal OTUs between A. glutinosa fine-root and mycorrhizal samples observed when all primers were combined showed no statistical significance (P = 0.45).
FIG. 2.
Average number of archaeal OTUs per sample type, obtained from five replicate samples. The fine-root (Fr; nonmycorrhizal) and mycorrhizal (Mm; inoculated with P. involutus) samples of the same tree are shown in pairs. The tree species of each sample type is given below the x axis. Error bars show standard errors of means. The occurrences of OTUs in fine-root and mycorrhizal samples of the same tree species are shown above the column pairs as statistically significant (**) or not significant (ns). P. sylvestris fine roots could not be tested for statistical significance. The patterns on the columns represent the primer combinations used (no pattern, combination 1; waves, combination 2; vertical stripes, combination 3).
The average number of archaeal OTUs in A. glutinosa samples (fine root and mycorrhizal combined) was significantly higher (average, 8.3 ± 1.85) than that in P. sylvestris (2.7 ± 0.97; two-tailed P = 0.02), P. abies (3.0 ± 0.76; two-tailed P = 0.02), or B. pendula (2.6 ± 0.88; two-tailed P = 0.02) samples. The archaeal OTU richness of deciduous trees (fine-root and mycorrhizal samples combined) did not differ significantly from that of coniferous trees (average two-tailed P = 0.06). However, deciduous (B. pendula and A. glutinosa) fine roots had statistically significantly higher levels of OTU richness than coniferous (P. sylvestris and P. abies) fine roots (average two-tailed P = 0.04), whereas the levels for deciduous mycorrhizas and coniferous mycorrhizas did not differ (average two-tailed P = 0.69). The average number of archaeal OTUs was statistically significantly higher in the fine-root (2.85 ± 1.05) and mycorrhizal (5.54 ± 0.83) samples than in the uncolonized humus (0.6 ± 0.4; P < 0.01 and P < 0.01, respectively) samples.
Phylogenetic analyses.
The majority of the 16S rRNA gene sequences obtained belonged to Crenarchaeota (Fig. 3). In addition, Euryarchaeota (Halobacterium) were found in two mycorrhizal samples (Fig. 4). The reproducibility of the DGGE method was tested by sequencing several OTUs with the same mobility from different samples. We found that although the OTUs showed similar mobility patterns in the DGGE run, the sequences were often quite different. According to DGGEs with PCR products obtained with primer combination 1, many of the fine-root OTUs were bacterial and were omitted from the study. However, all corresponding counterparts in the mycorrhizal samples were archaeal. Only two of the sequenced mycorrhizal OTUs were bacterial. Thus, all DGGE bands which were included in the analyses were with reasonable certainty archaeal.
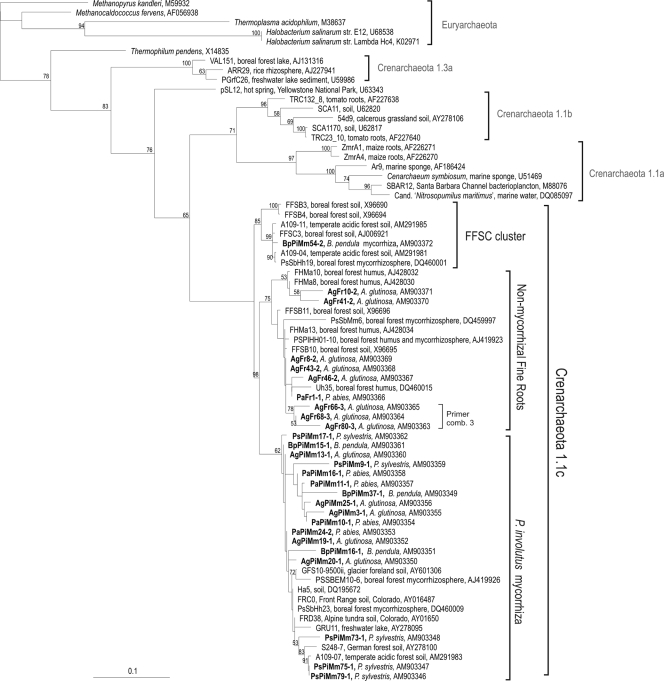
FIG. 3.
Maximum-likelihood phylogram presenting members of group 1.1c Crenarchaeota. The sequences found in this study are shown in bold script. The first two letters in the sample codes represent the tree species (Ps, P. sylvestris; Pa, P. abies; Bp, B. pendula; Ag, A. glutinosa). The middle two letters of the mycorrhizal sample codes represent the fungus (Pi, P. involutus). The last two letters represent the sample type (Fr, fine root; Mm, mycorrhiza). The numbers (1, 2, or 3) after the OTU code indicate the primer combination used to obtain the OTU. Values for nodes with over 50% bootstrap support are shown. The tree is rooted by Euryarchaeota.
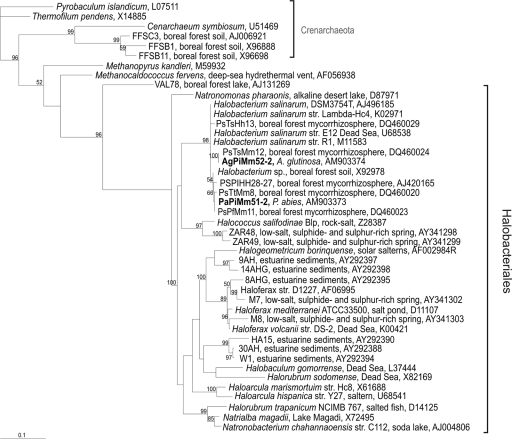
FIG. 4.
Maximum-likelihood phylogram presenting the Halobacteriales. The sequences of this study are shown in bold script, and the sequence codes are as described in the legend to Fig. 3. Values for nodes with over 50% bootstrap support are shown. The tree is rooted by Crenarchaeota.
A minimum of three bands from each OTU type from different sample types were chosen for sequencing. However, in most cases, only one sequence per OTU type with primer combination 1 was obtained due to the low storage stability of DGGE fragments. Nevertheless, two sequences of OTU type 16 were obtained (Fig. 1 and 3, sequences PaPiMm16-1 and BpPiMm16-1), and they fell into the same cluster. Corresponding fine-root OTU types also fell close to each other (AgFr66, -68, and -80; primer combination 3). Thus, within each sample type (fine root or mycorrhizal), the corresponding OTUs contained matching sequences, but sequences from similar OTUs from different sample types were not the same. The different primer combinations did not amplify the same archaeal 16S rRNA genes, and the sequences fell into different clusters in the phylogenetic analysis.
All the new crenarchaeotal 16S rRNA gene sequences fell into group 1.1c and clustered with Crenarchaeota previously found in boreal forest soil habitats (Fig. 3). The sequences were divided into three clusters, the FFSC cluster, the nonmycorrhizal fine-root cluster, and the P. involutus mycorrhiza cluster. The sequences from fine-root and mycorrhizal samples never fell into the same cluster. All sequences obtained from fine-root samples fell within the nonmycorrhizal fine-root cluster, with 16S rRNA gene sequences presently found only in Finnish forest soils. The A. glutinosa fine roots showed a unique community of group 1.1c Crenarchaeota. These sequences were not seen in any other samples, and they formed a separate cluster within the nonmycorrhizal fine-root cluster in the phylogenetic analyses (Fig. 3). All but one of the mycorrhizal sequences fell into the P. involutus mycorrhiza cluster. The P. involutus mycorrhiza cluster contained 16S rRNA gene sequences from different forest, field, and alpine soil and freshwater environments, covering temperate and boreal areas. BpPiMm54 was the only sequence associated with the FFSC cluster containing 1.1c Crenarchaeota from boreal and temperate forest soils.
The sequences of OTUs 5 (from P. sylvestris), 6 (P. sylvestris), 7 (P. sylvestris), 8 (P. sylvestris), 14 (A. glutinosa), 47 (B. pendula), 53 (B. pendula), and 55 (P. abies) from the mycorrhizal samples fell into the P. involutus mycorrhiza cluster, and the sequences of OTUs 2 (P. abies and B. pendula), 6 (B. pendula), and 71 (A. glutinosa) from the fine-root samples fell into the nonmycorrhizal fine-root cluster (Fig. 1). However, these sequences were too short for reasonable phylogenetic analysis. The 16S rRNA gene sequences for the two euryarchaeotal samples AgPiMm51 and AgPiMm52 fell with the Halobacterium, together with similar sequences from boreal forest P. sylvestris mycorrhizas.
DISCUSSION
The boreal forest soil habitats were again shown to harbor group 1.1c Crenarchaeota, although, generally, group 1.1b is the most common type found in soil (e.g., references 20 and 21). This is in accordance with the increasing number of studies reporting only or mostly 1.1c Crenarchaeota in boreal forest soil ecosystems (2, 3, 10, 12, 17, 40). A higher abundance of 1.1c Crenarchaeota than of 1.1b Crenarchaeota has also been shown to occur in soil from a temperate deciduous forest (13).
In our study, only three archaeal OTUs were obtained from soil currently uncolonized by mycorrhizal fungi. The sampled soil was not under the direct influence of the tree seedling and was not in contact with ectomycorrhizal hyphae, which distribute plant-derived substrates outside the immediate rhizosphere. We suggest that a rich vegetation cover with subsequent mycorrhizospheres plays an important role in the colonization of soil by 1.1c Crenarchaeota. This also agrees with the results obtained by Nicol et al. (20), who showed that in glacier foreland soil, no 1.1c Crenarchaeota were found until the soil had been ice free for at least 135 years, had a stable vegetation cover, and had a well-developed organic top layer. Our theory is supported by the fact that recently uncovered soil is primarily colonized by weakly to moderately mycotrophic plant species but that the mature flora mostly represent highly mycotrophic plant species (27). These plants are colonized by mycorrhizal fungi, and selection pressure on rhizosphere bacteria has been shown to be induced by these fungi (1, 15, 33). These more organic soils are also likely to contain more saprophytic fungi than the young mineral soils. Currently, there is no information about the relationship between saprophytic fungi and archaea. We also presume that mycorrhizal fungi influenced the archaeal phylotypes retrieved by Oline et al. (22), who sampled 1-cm-diameter and 10-cm-deep cores of mature, vegetated soils, most of which were dominated by ectomycorrhizal plant species, such as the tree species Pinus ponderosa, Pinus contorta, and Pseudotsuga menziesii. Since root and hyphal materials were not completely excluded from the samples, it is likely that mycorrhizas or mycorrhizal hyphae ended up in the soil samples when the soil was sieved with a 2-mm mesh. Kemnitz et al. (13) found that in temperate deciduous forests, 1.1c Crenarchaeota are also dominated by heavily mycorrhizal plants. The crenarchaeotal 1.1c 16S rRNA gene sequences found in mycorrhizal samples in our study were similar to the sequences retrieved from soils in the above-mentioned studies (13, 22). Only Sliwinsky and Goodman (34) did not find group 1.1c Crenarchaeota in soils vegetated by commonly mycorrhizal plants. Wallander et al. (38) have calculated that boreal forest humus contains a substantial amount of ectomycorrhizal hyphae, between 125 and 200 kg ha−1, in the organic top layer, equaling 1.25 to 2 g cm−2. Levels for all ectomycorrhizal mycelia (including ectomycorrhizal mantle structures) were estimated to be as high as 700 to 900 kg ha−1. This shows that ectomycorrhizal fungi constitute a huge part of the biomass in the forest soil associated with trees.
There was a great difference in archaeal community between the rhizospheres of some of the tree species, even when they were grown in soil originating from the same location. This contrasts with Sliwinski and Goodman's results (34), where significant differences were detected not in the rhizospheres of plants from the same site but only between different geographical locations. Our trees were all grown in the same humus, obtained from a dry pine forest. The differences in archaeal rhizosphere communities would certainly be even greater if the tree species had been grown in soil from a forest dominated by the tree in question, but then we would have been in the same situation as Sliwinski and Goodman (34), where the influence of the plant could not be separated from that of the location. Nicol et al. (20) studied the rhizospheres of different plant species in Austrian alpine glacier front soil. Minor differences were observed in the community structures of archaea associated with the roots of different plant species, but only between plants from soils of different ages. These results could be explained by the mycorrhizosphere effect, since few natural root systems are devoid of mycorrhizal fungi. In our present study, we found that the influence of the tree on the archaeal populations was minimized by the impact of the symbiotic ectomycorrhizal fungus colonizing the tree roots. Mycorrhizal fine roots had more even frequencies of archaeal OTUs, and these sequences were more similar to each other than to the sequences from uncolonized fine roots.
The mycorrhizal roots of most tree species harbored more-diverse populations of archaea than uncolonized fine roots or humus. Simon et al. (32) estimated that the doubling time of group 1.1b Crenarchaeota inhabiting tomato roots was 8 days. If the group 1.1c Crenarchaeota detected in this study grow equally slowly, mycorrhizas may present a more constant environment for these organisms than the rapidly growing root tips. It is also possible that the archaea are simply outcompeted by more-opportunistic bacteria on the root surfaces. This could explain the relatively high number of bacterial 16S rRNA gene sequences obtained from the fine-root samples. The crenarchaeotal 16S rRNA gene sequences obtained from fine roots formed a distinct cluster, separate from the mycorrhizal clusters. Interestingly, their closest neighbors were crenarchaeotal 16S rRNA gene sequences thus far found only in Finnish dry pine forest soil habitats (soil, mycorrhizospheres, and tree roots). In contrast to the crenarchaeotal 16S rRNA gene sequences detected in the mycorrhizal samples, none of those detected in the fine-root samples were similar to 16S rRNA gene sequences from other locations or habitat types.
There was no statistically significant difference in the occurrence of archaeal OTUs in the mycorrhizal samples derived from the four different tree species, and the sequences of the archaeal 16S rRNA gene OTUs all fell within the same cluster. This confirms that the mycorrhizal fungus in the roots attracts its own archaeal community independent of the one colonizing the rhizosphere, which is in agreement with our earlier results (3) showing that the archaeal population in the P. sylvestris mycorrhizosphere varies with the species of ectomycorrhizal fungus colonizing the roots. Euryarchaeota were detected in only one P. abies and one A. glutinosa mycorrhizal sample. These findings confirm our earlier results showing that, unlike some other ectomycorrhizal fungi, P. involutus does not particularly attract Euryarchaeota (3).
With the exception of A. glutinosa fine roots, the numbers of archaeal OTUs were low, and the archaeal 16S rRNA gene fragments detected appeared to be similar in the fine-root samples of the different trees. This is slightly surprising, as the different tree species influence the soil in their root and mycorrhizal vicinities by different means (26). The pH of the soil in a coniferous forest is low, and the typical pH of Finnish dry pine forest soils is between 4 and 5. Conifers, especially spruce, may decrease the pH of the soil to as low as 3.5, whereas birch has been shown to increase the soil pH up to 5.7 and enhance the cycling of nutrients (16). Deciduous litter also contains more easily leachable carbon, whereas coniferous litter is acidic and contains recalcitrant material, such as waxes, phenolics, and lignin (9, 24). The litter was not a factor in this study, because our experiment stretched over only one growing season. However, the short time of this experiment showed that A. glutinosa seedlings promoted both different numbers and different types of Crenarchaeota on their fine roots.
The A. glutinosa fine-root samples displayed the highest overall number of different archaeal OTUs in this study, followed by the A. glutinosa mycorrhizal samples (Fig. 2). The A. glutinosa fine-root samples were also the only ones that presented PCR products with primer combination 3 which formed a separate cluster within the nonmycorrhizal fine-root cluster. A. glutinosa is known for its ability to attract nodule-forming, nitrogen-fixing bacteria. No nodules were discovered on the roots of our test trees, but the A. glutinosa rhizospheric environment and consequently its microbial flora are probably specialized and different from the other tree rhizospheric microbial communities. This is evident also in the residing archaeal population. With the exception of the A. glutinosa fine-root results, archaea did not appear to thrive on tree fine roots but, in agreement with our earlier studies, preferred the mycorrhizas (2, 3). The primer set used had a great effect on the detection of archaea in the various samples. This is a known problem in studying natural, uncultured populations where even slight adjustments of procedures may alter the results. The lack of redundancy between the OTUs produced with the different primer combinations was conspicuous. However, the outer primers of primer combination 1 targeted a broad spectrum of microbial 16S rRNA genes and appeared to select for the Crenarchaeota in the P. involutus mycorrhiza cluster (Fig. 3), while the more archaea-specific outer primers selected for Crenarchaeota affiliating with the nonmycorrhizal fine-root cluster. It is possible that new primer combinations may reveal yet undiscovered archaeal groups in fine root, mycorrhizal, and uncolonized humus habitats.
Our conclusion is that different tree species have different influences on soil archaeal communities. These differences are diminished, however, by the ectomycorrhizal fungus colonizing the roots. The ectomycorrhizal fungus P. involutus attracts group 1.1c Crenarchaeota, and the crenarchaeotal populations in the mycorrhizas are different from those in the uncolonized fine roots of boreal forest trees.
Acknowledgments
This work was supported by the Maj and Tor Nessling Foundation, Svenska Kulturfonden, and the University of Helsinki.
Graeme Nicol and Leone Montonen are warmly thanked for their critical comments on the manuscript. Uwe Münster and Jari Valkonen are gratefully acknowledged for their support during this work.
Footnotes
Published ahead of print on 31 October 2008.
REFERENCES
- 1.Artursson, V., R. D. Finlay, and J. K. Jansson. 2005. Combined bromodeoxyuridine immunocapture and terminal-restriction fragment length polymorphism analysis highlights differences in the active soil bacterial metagenome due to Glomus mosseae inoculation or plant species. Environ. Microbiol. 7:1952-1966. [DOI] [PubMed] [Google Scholar]
- 2.Bomberg, M., G. Jurgens, A. Saano, R. Sen, and S. Timonen. 2003. Nested PCR detection of archaea in defined compartments of pine mycorrhizospheres developed in boreal forest humus microcosms. FEMS Microbiol. Ecol. 43:163-171. [DOI] [PubMed] [Google Scholar]
- 3.Bomberg, M., and S. Timonen. 2007. Distribution of Cren- and Euryarchaeota in Scots pine mycorrhizospheres and boreal forest humus. Microb. Ecol. 54:406-416. [DOI] [PubMed] [Google Scholar]
- 4.Buckley, D. H., J. R. Graber, and T. M. Schmidt. 1998. Phylogenetic analysis of nonthermophilic members of the kingdom Crenarchaeota and their diversity and abundance in soils. Appl. Environ. Microbiol. 64:4333-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chelius, M. K., and E. W. Triplett. 2001. The diversity of Archaea and Bacteria in association with the roots of Zea mays L. Microb. Ecol. 41:252-263. [DOI] [PubMed] [Google Scholar]
- 6.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLong, E. F. 1998. Everything in moderation: archaea as ‘non-extremophiles’. Curr. Opin. Genet. Dev. 8:649-654. [DOI] [PubMed] [Google Scholar]
- 8.Groβkopf, R., S. Stubner, and W. Liesack. 1998. Novel euryarchaeotal lineages detected on rice roots and in the anoxic bulk soil of flooded rice microcosms. Appl. Environ. Microbiol. 64:4983-4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris, M. M., and L. O. Safford. 1996. Effect of season and four tree species on soluble carbon content in fresh and decomposing litter of temperate forests. Soil Sci. 161:130-135. [Google Scholar]
- 10.Jurgens, G., and A. Saano. 1999. Diversity of soil Archaea in boreal forest before, and after clear-cutting and prescribed burning. FEMS Microbiol. Ecol. 29:205-213. [Google Scholar]
- 11.Jurgens, G., F. Glöckner, R. Amann, A. Saano, L. Montonen, M. Likolammi, and U. Münster. 2000. Identification of novel Archaea in bacterioplankton of a boreal forest lake by phylogenetic analysis and fluorescent in situ hybridization. FEMS Microbiol. Ecol. 34:45-56. [DOI] [PubMed] [Google Scholar]
- 12.Jurgens, G., K. Lindström, and A. Saano. 1997. Novel group within the kingdom Crenarchaeota from boreal forest soil. Appl. Environ. Microbiol. 63:803-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kemnitz, D., S. Kolb, and R. Conrad. 2007. High abundance of Crenarchaeota in a temperate acidic forest soil. FEMS Microbiol. Ecol. 60:442-448. [DOI] [PubMed] [Google Scholar]
- 14.Könneke, M., A. E. Bernhard, J. R. de la Torre, C. B. Walker, J. B. Waterbury, and D. A. Stahl. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543-546. [DOI] [PubMed] [Google Scholar]
- 15.Marschner, P., and S. Timonen. 2005. Bacterial community composition and activity in rhizospheres of roots colonised by arbuscular mycorrhizal fungi, p. 139-149. In K. G. Mukerji, C. Manoharachary, and J. Singh (ed.), Microbial activity in the rhizosphere. Springer-Verlag, Berlin, Germany.
- 16.Mikola, P. 1985. The effect of tree species on the biological properties of forest soil. Naturvårdsverkets rapport 3017. Statens Naturvårdsverk, Solna, Sweden.
- 17.Nicol, G. W., C. D. Campbell, S. J. Chapman, and J. I. Prosser. 2007. Afforestation of moorland leads to changes in crenarchaeal community structure. FEMS Microbiol. Ecol. 60:51-59. [DOI] [PubMed] [Google Scholar]
- 18.Nicol, G. W., L. A. Glover, and J. I. Prosser. 2003. Spatial analysis of archaeal community structure in grassland soil. Appl. Environ. Microbiol. 69:7420-7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicol, G. W., D. Tscherko, L. Chang, U. Hammesfahr, and J. I. Prosser. 2006. Crenarchaeal community assembly and microdiversity in developing soils at two sites associated with deglaciation. Environ. Microbiol. 8:1382-1393. [DOI] [PubMed] [Google Scholar]
- 20.Nicol, G. W., D. Tscherko, T. M. Embley, and J. I. Prosser. 2005. Primary succession of soil Crenarchaeota across a receding glacier foreland. Environ. Microbiol. 7:337-347. [DOI] [PubMed] [Google Scholar]
- 21.Ochsenreiter, T., D. Selezi, A. Quaiser, L. Bonch-Osmolovskaya, and C. Schleper. 2003. Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ. Microbiol. 5:787-797. [DOI] [PubMed] [Google Scholar]
- 22.Oline, D. K., S. K. Schmidt, and M. C. Grant. 2006. Biogeography and landscape-scale diversity of the dominant Crenarchaeota of soil. Microb. Ecol. 52:480-490. [DOI] [PubMed] [Google Scholar]
- 23.Peters, S., S. Koschinsky, F. Schwieger, and C. C. Tebbe. 2000. Succession of microbial communities during hot composting as detected by PCR-single-strand-conformation polymorphism-based genetic profiles of small-subunit rRNA genes. Appl. Environ. Microbiol. 66:930-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prescott, C. E., L. L. Blevins, and C. Staley. 2004. Litter decomposition in British Columbia forests: controlling factors and influences of forestry activities. BC J. Ecosyst. Manage. 5:44-57. [Google Scholar]
- 25.Preston, C. M., K. Y. Wu, T. F. Molinski, and E. F. DeLong. 1996. A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov., sp. nov. Proc. Natl. Acad. Sci. USA 93:6241-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Priha, O., and A. Smolander. 1997. Microbial biomass and activity in soil and litter under Pinus sylvestris, Picea abies and Betula pendula at originally similar field afforestation sites. Biol. Fertil. Soils 24:45-54. [Google Scholar]
- 27.Read, D. J., and K. H. Wandter. 1980. Observations on the mycorrhizal status of some alpine plant communities. New Phytol. 88:341-352. [Google Scholar]
- 28.Rincon, B., F. Raposo, R. Borja, J. M. Gonzalez, M. C. Portillo, and C. Saiz-Jimenez. 2006. Performance and microbial communities of a continuous stirred tank anaerobic reactor treating two-phases olive mill solid wastes at low organic loading rates. J. Biotechnol. 121:534-543. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Sandaa, R. A., O. Enger, and V. Torsvik. 1999. Abundance and diversity of Archaea in heavy-metal-contaminated soils. Appl. Environ. Microbiol. 65:3293-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simon, H. M., J. A. Dodsworth, and R. M. Goodman. 2000. Crenarchaeota colonize terrestrial plant roots. Environ. Microbiol. 2:495-505. [DOI] [PubMed] [Google Scholar]
- 32.Simon, H. M., C. E. Jahn, L. T. Bergerud, M. K. Sliwinski, P. J. Weimer, D. K. Willis, and R. M. Goodman. 2005. Cultivation of mesophilic soil crenarchaeotes in enrichment cultures from plant roots. Appl. Environ. Microbiol. 71:4751-4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh, B. K., N. Nunan, K. P. Ridgway, J. McNicol, J. P. W. Young, T. J. Daniell, J. I. Prosser, and P. Millard. 2008. Relationship between assemblages of mycorrhizal fungi and bacteria on grass roots. Environ. Microbiol. 10:534-541. [DOI] [PubMed] [Google Scholar]
- 34.Sliwinski, M. K., and R. M. Goodman. 2004. Comparison of crenarchaeal consortia inhabiting the rhizosphere of diverse terrestrial plants with those in bulk soil in native environments. Appl. Environ. Microbiol. 70:1821-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes in bacterial systematics. p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics, 1st ed. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 36.Timonen, S., R. D. Finlay, B. Söderström, and M. Raudaskoski. 1993. Identification of cytosceletal components in pine ectomycorrhizas. New Phytol. 124:83-92. [Google Scholar]
- 37.Timonen, S., K. S. Jørgensen, K. Haahtela, and R. Sen. 1998. Bacterial community structure at defined locations of Pinus sylvestris Suillus bovinus and Pinus sylvestris Paxillus involutus mycorrhizospheres in dry pine forest humus and nursery peat. Can. J. Microbiol. 44:499-513. [Google Scholar]
- 38.Wallander, H., L. O. Nilsson, D. Hagerberg, and E. Bååth. 2001. Estimation of the biomass and seasonal growth of external mycelium of ectomycorrhizal fungi in the field. New Phytol. 151:753-760. [DOI] [PubMed] [Google Scholar]
- 39.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yrjälä, K., R. Katainen, G. Jurgens, U. Saarela, A. Saano, M. Romantchuk, and H. Fritze. 2004. Wood ash fertilization alters the forest humus Archaea community. Soil Biol. Biochem. 36:199-201. [Google Scholar]