FIG. 1.
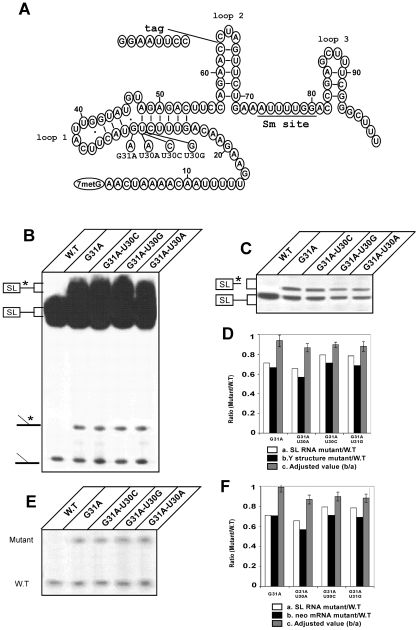
Mutated SL RNAs of L. collosoma that cannot undergo pseudouridylation are affected only slightly as trans-splicing substrates. (A) Sequences and secondary structures of L. collosoma SL RNA and its mutated derivatives. The loops, Sm site, intron tag, G31A substitution, and U30 substitutions are indicated. (B) Effects of U30 mutations on the first step of trans splicing. Total RNA from the indicated cell lines was primer extended with SL RNA oligonucleotide 9825. The cDNA products were separated on a denaturing 6% polyacrylamide gel. The 5′ end of the SL RNA is indicated. The positions of the cDNAs derived from full-length mutated SL RNA and its corresponding Y structure are marked with asterisks. (C) Northern analysis of SL RNAs bearing U30 mutations. Total RNA was separated on a 10% polyacrylamide denaturing gel and electroblotted onto a nylon membrane. The identities of the mutations are indicated at the top. The blot was hybridized with oligonucleotide 9825, which recognizes both the wild-type (W.T) and the mutated SL RNAs. (D) Quantitative analysis of utilization of U30 SL RNA mutations in the first step of splicing. SL RNA levels (mutated and wild type) were estimated by densitometry. (a) Ratio between the levels of mutated and wild-type SL RNAs, calculated from the data presented in Fig. 1B and C. (b) Ratio between the levels of the mutated and wild-type Y structures, calculated from the data presented in Fig. 1B. (c) Adjusted values accounting for differential expression, i.e., the Y-structure ratio divided by the SL RNA ratio. Error bars indicate standard deviations of the adjusted values (n = 5; P < 0.01) for all cases. (E) Effects of U30 mutations on the second step of trans splicing. Total RNA from the indicated cell lines was reverse transcribed with an end-labeled primer (36815), as described previously (41) and in Materials and Methods. The primer extension products were separated on a denaturing 6% polyacrylamide gel. The diagnostic cDNAs derived from the presence of mutated or wild-type SL RNA exons are indicated. (F) Quantitative analysis of utilization of U30 SL RNA mutations in the second step of trans splicing. The level of mature neo mRNA was estimated using densitometry. (a) Ratio between the levels of mutated and wild-type SL RNAs, calculated from the Northern analysis data presented in Fig. 1C. (b) Ratio between the levels of mutated and wild-type SL RNA stops, calculated from the data presented in Fig. 1E. (c) Adjusted values obtained by dividing the SL RNA stop ratio by the SL RNA ratio. Error bars indicate standard deviations of the adjusted values (n = 5; P < 0.01) for all cases.