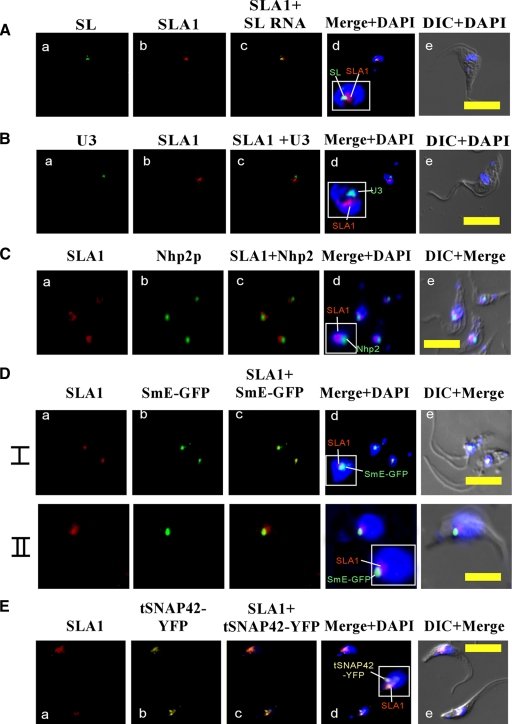
FIG. 3.
Localization of SLA1 in the nucleus. (A) Colocalization of SLA1 and SL RNA. (a) In situ hybridization with DIG-labeled antisense SL RNA probe (green); (b) in situ hybridization with biotin-labeled antisense SLA1 probe (red); (c) merge of a and b; (d) nuclear staining with DAPI merged with a and b; (e) differential interference contrast (DIC) merged with nuclear staining using DAPI. (B) Localization of SLA1 relative to the U3 snoRNA. (a) In situ hybridization of DIG-labeled U3 RNA probe (green); (b) in situ hybridization of biotin-labeled SLA1 antisense probe (red); (c) merge of a and b; (d) nuclear staining with DAPI merged with a and b; (e) DIC merged with nuclear staining with DAPI. (C) Localization of SLA1 relative to Nhp2p. (a) In situ hybridization with biotin-labeled antisense SLA1 RNA probe (red); (b) immunofluorescence with anti-Nhp2 antibody (green) (the antibodies [31] were diluted 1:200); (c) merge of a and b; (d) nuclear staining with DAPI merged with a and b; (e) DIC merge of a, b, and d. (D) Localization of SLA1 relative to SmE-GFP. Cells expressing the SmE-GFP construct (31) were fixed with 4% (vol/vol) formaldehyde and hybridized with labeled RNA probes (I and II represent two fields of the same experiment). (a) In situ hybridization with biotin-labeled antisense SLA1 RNA probe (red); (b) fluorescence of SmE-GFP (green); (c) merge of a and b; (d) nuclear staining with DAPI merged with a and b; (e) DIC merge of a, b, and d. (E) Localization of SLA1 relative to tSNAP42-YFP. Cells (see Materials and Methods) were fixed with 4% (vol/vol) formaldehyde and hybridized with labeled RNA probes. (a) In situ hybridization with biotin-labeled antisense SLA1 RNA probe (red); (b) fluorescence of tSNAP42-YFP (yellow); (c) merge of a and b; (d) nuclear staining with DAPI merged with a and b; (e) DIC merge of a, b, and d. All slides were observed under a Zeiss LSM 510 META inverted microscope. Bar, 10 μm.