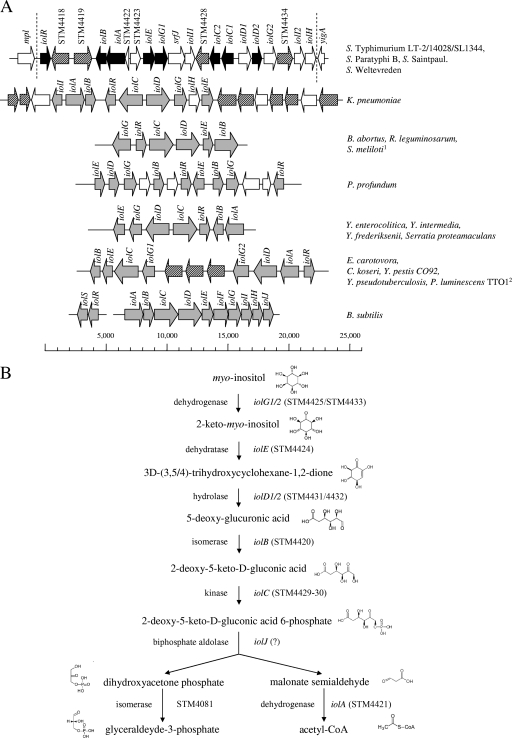
FIG. 1.
(A) Examples of iol divergons. GEI 4417/4436 (22.6 kb) of S. enterica serovar Typhimurium is presented in comparison to the structural organization of iol genes from B. subtilis and several gram-negative bacteria. Salmonella genes experimentally demonstrated in this study to belong to the inositol divergon are depicted in black; their homologues in other pathogens are shown in gray. Genes encoding putative permeases are hatched. 1, in this organism, iolG is transcribed in the same orientation as the other genes; 2, the iol cluster of P. luminescens is similar to that of E. carotovora but lacks two of three putative permease genes. (B) Reconstruction of the pathway for MI degradation in S. enterica serovar Typhimurium. Seven stepwise reactions are involved in MI degradation to glyceraldehyde-3-phosphate and acetyl coenzyme A (acteyl-CoA). None of the genes from GEI 4417/4436 encodes a homologue of a biphosphate aldolase. Chemical structures were taken from the Kyoto encyclopedia of genes and genomes (13).