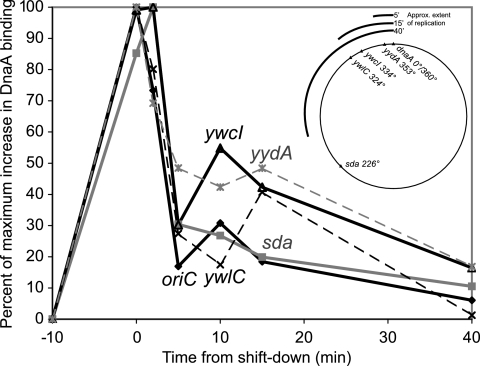
FIG. 4.
Release of DnaA binding after resumption of replication. dnaD(Ts) cells (KPL73) in exponential growth phase were shifted to the restrictive temperature for 60 min and then shifted back to the permissive temperature in order to generate a synchronous round of replication initiation. Samples were taken for ChIP-PCR analysis immediately before and at the end of the incubation at the restrictive temperature and 2, 5, 10, 15, and 40 min after the shift back to the permissive temperature. DnaA binding was analyzed at oriC/dnaA (solid diamonds and black lines), yydA (gray asterisks and gray dashed lines), ywcI (solid triangles and black lines), ywlC (black X characters and black dashed lines), and sda (filled gray squares and solid gray lines). Data were normalized according to the basal and maximal levels of signal detected to facilitate comparison of binding kinetics at sites with different levels of DnaA binding (see, for example, levels at dnaA/oriC and sda in Fig. 2). Data are shown in the vicinities of mntH (A), yloB (B), pksN (C), nrdE (D), sunA (E), yrhC (F), ywpH (G), spo0J (H), and ysmB (I). The map in the upper right shows the position of each analyzed site on the chromosome and the approximate extent of replication 5, 15, and 40 min after release.