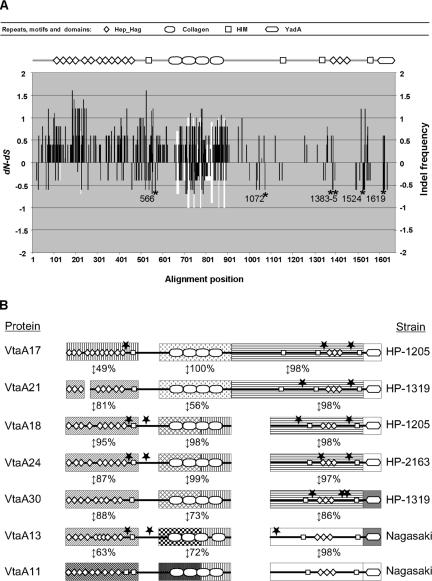
FIG. 5.
Selective pressure of vtaA10 orthologs and mosaic structure of VtaA molecules. (A) A multiple alignment of vtaA17, vtaA27, vtaA23, and vtaA25 was performed for selective pressure determination. The dN − dS for each codon position of the vtaA10 strict homologs is indicated by the black bars and the frequency of indels by the white bars. The alignment position axis is given in amino acid numbers. For the whole molecule, the mean dN − dS was 0.599163 with a log likelihood of −10,670.5. Six positions (indicated by asterisks) were detected as negatively selected by the SLAC codon-based maximum likelihood method (P < 0.05). The protein architecture of VtaA10 is represented at the top of the figure; a positive selective pressure applies to the part of the gene coding for the N-terminal Hep_Hag and HIM repeats and motifs, respectively. (B) To illustrate the mosaic structure of a sample of VtaA molecules, modules of the passenger domain were selected and compared using BlastP. Pairwise amino acid identities are indicated as percentages. Potential recombination breakpoints detected by more than five of the seven algorithms included in the RDP2 package with statistical significance (P < 0.05) are indicated by stars. Modules and connective regions are not to scale, and blanks indicate indels.