Abstract
The present study was designed to characterize phenotypically and genotypically seven Arcanobacterium haemolyticum strains obtained from infections of six horses. All seven strains showed the cultural and biochemical properties typical of A. haemolyticum and were susceptible to most of the antibiotics tested. The species identification could be confirmed by amplification and sequencing of the 16S rRNA gene and the 16S-23S rRNA intergenic spacer region and by PCR amplification of species-specific parts of the gene encoding phospholipase D in A. haemolyticum. Use of the latter could possibly improve future identification of this generally human pathogenic bacterial species which, according to the present results, seems to occur also in infections of horses.
Arcanobacterium haemolyticum, originally known as Corynebacterium haemolyticum, was first described in 1946 and reported to be the cause of exudative pharyngitis and skin infections in humans (23). This bacterial species, formerly considered to be a mutant form of Actinomyces (Corynebacterium) pyogenes (1), has been reclassified as a single species in the newly proposed genus Arcanobacterium (8). In the following years, the mainly animal pathogenic species Actinomyces pyogenes and human pathogenic Actinomyces bernardiae were renamed Arcanobacterium pyogenes and Arcanobacterium bernardiae, respectively, and five new species, namely, Arcanobacterium bialowiezense, Arcanobacterium bonasi, Arcanobacterium hippocoleae, Arcanobacterium phocae, and Arcanobacterium pluranimalium, were assigned to this genus (14, 19, 20, 28). A. haemolyticum, known as a pathogen causing nonstreptococcal pharyngitis (6, 22), could also be isolated from systemic and deep-seated infections of humans. The latter was reviewed by Skov et al. (31) and Tan et al. (33). However, the isolation of A. haemolyticum from animals appears to be rare. Tyrrell et al. (34) discussed the etiological role of a single A. haemolyticum strain isolated from a periodontal infection of a rabbit.
The aim of the present study was to characterize phenotypically and genotypically seven A. haemolyticum strains isolated from infections of six horses over a period of 10 years.
MATERIALS AND METHODS
Bacterial cultures.
A total of 21 bacterial cultures were used in this study. The cultures included the reference strains A. haemolyticum DSM 20595, A. bernardiae DSM 9152, A. bialowiezense DSM 17162, A. bonasi DSM 17163, A. hippocoleae DSM 15539, A. phocae DSM 10002, A. pluranimalium DSM 13483, A. pyogenes DSM 20630, Corynebacterium pseudotuberculosis biovar equi DSM 7177, and C. pseudotuberculosis biovar ovis DSM 7179 and seven bacterial cultures isolated from infections of six horses which were presumptively identified as A. haemolyticum. Further data about the origins of the seven strains are summarized in Table 1.
TABLE 1.
Origin of the seven A. haemolyticum strains isolated from six horses
| Horse | Date of isolation (day.mo.yr) | Origin | A. haemolyticum strain designation | Other isolated microorganism(s) |
|---|---|---|---|---|
| 1 | 15.07.1997 | Wound infection; surgical clinic | 3801 | Streptococcus equi subsp. zooepidemicus, coliform bacteria, Staphylococcus aureus, Escherichia coli, Proteus spp. |
| 2 | 05.08.1997 | Wound infection; surgical clinic | 4205 | S. equi subsp. zooepidemicus, S. aureus, E. coli, Proteus spp., Corynebacterium spp. |
| 3 | 10.12.1997 | Wound infection; surgical clinic | 6879 | Proteus spp., S. aureus, E. coli, aerobic Bacillus spp., Morganella morganii |
| 4 | 17.02.2003 | Fistula after castration; surgical clinic | 708 | S. equi subsp. zooepidemicus, Corynebacterium spp., Pasteurella caballi, Fusobacterium nucleatum |
| 5 | 2005 | Dermatitis; no further data available | P664 | No data available |
| 6 | 09.05.2007 | Dermatitis; veterinarian | P3333 | Proteus spp., alpha-hemolytic streptococci, Citrobacter koseri, S. equi subsp. zooepidemicus, S. aureus, Pasteurella caballi, Actinobacillus equuli |
| 6 | 09.05.2007 | Scurf; veterinarian | P3334 | S. aureus, alpha-hemolytic streptococci, Corynebacterium spp., S. equi subsp. zooepidemicus |
Phenotypic properties.
Hemolytic properties of the bacteria were examined on blood agar containing 5% sheep or rabbit blood after incubation of the bacteria for 24 to 48 h in a candle jar. The rabbit blood was obtained from Harlan Winkelmann, Borchen, Germany. In addition, a CAMP-like test was performed on sheep blood agar with Streptococcus agalactiae and Rhodococcus equi reference cultures, and the reverse CAMP test was performed with a beta-hemolytic Staphylococcus aureus strain (18, 21). The indicator microorganisms were obtained from the strain collection of Institut für Pharmakologie und Toxikologie. The A. haemolyticum cultures were additionally cultivated on Loeffler agar (2) and for determination of cross-reactions with streptococcal serogroup G-specific antisera investigated with a commercial grouping kit (streptococcal identification kit; Oxoid, Wesel, Germany). For biochemical characterization, the API Coryne test system (Biomerieux, Nürtingen, Germany) was used according to the instructions of the manufacturer as described previously (10). In addition, tablets containing substrates for investigation of the enzymes alkaline phosphatase, α-mannosidase, and pyrrolidonyl arylamidase (Inverness Medical, Köln, Germany) were used according to the instructions of the manufacturer (Rosco Diagnostica A/S, Taastrup, Denmark). Further enzyme studies were performed using the substrates 4-methylumbelliferyl-β-d-galactopyranoside (5 μmol), 4-methylumbelliferyl-β-d-glucuronide (15 μmol), and 4-methylumbelliferyl-N-acetyl-β-d-glucosaminide (5 μmol) (Sigma, Steinheim, Germany) dissolved in 0.2 ml of dimethyl sulfoxide as described by Maddocks and Greenan (24). The individual stock solutions were diluted to 10 ml in acetate buffer, pH 5.2 (32). The enzyme assay was performed by the method of Slifkin and Gil (32). The bacteria were additionally investigated for catalase activity with 3% H2O2 on microscopic slides for hyaluronidase enzyme activity with a Streptococcus equi subsp. equi reference strain by the method of Raus and Love (29) for DNase activity on DNase test agar (Merck, Darmstadt, Germany) and for acetoin production with methyl red-Voges-Proskauer broth (Merck).
Antibiotic resistance.
The bacterial strains were studied for antimicrobial susceptibility on Mueller-Hinton agar (Oxoid) plates containing 5% sheep blood using enrofloxacin (5 μg, Oxoid), erythromycin (15 μg), gentamicin (10 μg), penicillin G (10 IU), tetracycline (30 μg), and sulfamethoxazole-trimethoprim (23.75 μg of sulfamethoxazole and 1.25 μg for trimethoprim) disks (Mast Diagnostics, Reinfeld, Germany). The results were recorded according to NCCLS criteria (26) after 48 h of incubation at 37°C in a candle jar by measuring the antimicrobial zone diameters.
Genotypic properties.
For DNA extraction, a single colony of each isolate was cultivated and incubated for 48 h on sheep blood agar in a candle jar. Five to 10 colonies of the freshly subcultured strain were subsequently suspended in 180 μl TE buffer (10 mmol/liter Tris-HCl, 1 mmol/liter EDTA [pH 7.2]) containing 5 μl mutanolysin (10 U/μl; Sigma). After incubation for 1 h at 37°C, 25 μl proteinase K (Qiagen, Hilden, Germany) and 200 μl AL lysis buffer (Qiagen) were added, and the suspension was incubated for an additional 2 h at 56°C. The DNA was subsequently isolated by using a DNeasy tissue kit according to the manufacturer's instructions (Qiagen).
Sequencing of 16S rRNA gene and 16S-23S rRNA ISR.
The 16S rRNA gene of the seven A. haemolyticum strains was amplified with an expected size of 1,403 bp by using the oligonucleotide primers 16SUNI-L (5′-AGA GTT TGA TCA TGG CTC AG-3′) and 16SUNI-R (5′-GTG TGA CGG GCG GTG TGT AC-3′) (16), which corresponded to bases 8 to 27 and to bases 1391 to 1410 of the 16S rRNA gene sequence of Escherichia coli (NCBI accession number J01859), respectively. The intergenic spacer region (ISR) was amplified with an expected size of 600 bp by using the oligonucleotide primers c (5′-TTG TAC ACA CCG CCC GTC A-3′) and b (5′-GGT ACC TTA GAT GTT TCA GTT C-3′) described by Kostman et al. (15) and Chanter et al. (7).
Both PCR amplifications were performed with the following reaction mixture (30 μl): 1 μl (10 pmol/μl) of each primer, 0.6 μl (10 mmol/liter) of deoxynucleoside triphosphates (Fermentas, St. Leon-Rot, Germany), 3 μl GeneAmp 10× PCR Gold buffer (150 mM Tris-HCl, 500 mM KCl [pH 8.0]) (Applied Biosystems, Darmstadt, Germany), 1.8 μl MgCl2 (25 mM) (Applied Biosystems), 0.2 μl AmpliTaq Gold polymerase (5 U/μl, Applied Biosystems), and 19.9 μl sterile aqua dest. Finally, 2.5 μl DNA template was added to this reaction mixture. The PCR program for the 16S rRNA gene was carried out as follows: one step of 10 min at 95°C; 30 cycles, with 1 cycle consisting of 30 s at 95°C, 60 s at 58°C, and 60 s at 72°C; and one step of 7 min at 72°C. For ISR, this program was followed by one step of 10 min at 95°C; 30 cycles, with 1 cycle consisting of 70 s at 95°C, 70 s at 45°C, and 70 s at 72°C; and one step of 7 min at 72°C using a Biometra T3000 thermocycler (Biometra, Göttingen, Germany) or Gene Amp PCR system 2400 (Perkin-Elmer, Rodgau Jügesheim, Germany). The PCR products (8 μl) were mixed with 2 μl loading dye solution (Fermentas) and separated by 2% agarose gel electrophoresis (Biozym, Hessisch-Oldendorf, Germany) at 120 V in 1× TAE buffer (4.0 mmol/liter Tris-HCl, 1 mmol/liter EDTA [pH 8.0]) with a 100-bp DNA ladder (Roche, Mannheim, Germany) as the molecular size standard. The PCR products were than stained for 5 min with 5 μl/ml ethidium bromide solution (Sigma). The amplicons were then visualized under a UV trans-illuminator (Bio-Rad, München, Germany) or ImageMaster VDS (Pharmacia Biotech, Freiburg, Germany).
In parallel, the PCR products were purified using a commercial PCR purification kit (QIAquick PCR purification kit) as recommended by the manufacturer (Qiagen). The purified DNA was sequenced by SEQLAB Sequence Laboratories (Göttingen, Germany). The obtained sequences of the 16S rRNA gene and of ISR of the seven A. haemolyticum strains were aligned, further analyzed using the cluster method of the MegAlign program (DNASTAR Inc., Madison, WI), and compared with the nucleotide sequences of the 16S rRNA gene and ISR of all eight Arcanobacterium reference strains obtained from NCBI GenBank: A. haemolyticum (GenBank accession numbers AJ234059 and EU194564, respectively), A. bernardiae (X79224; EU194562), A. bialowiezense (AJ879696; EU194569), A. bonasi (AJ879697; EU194570), A. hippocoleae (AJ300767; EU194568), A. phocae (X97049; EU194566), A. pluranimalium (AJ250959; EU194567), and A. pyogenes (X79225; EU194563).
Design of an A. haemolyticum phospholipase D gene-specific PCR.
The A. haemolyticum phospholipase D gene (pld)-specific oligonucleotide primers were designed by OLIGO 4 primer analysis software (version 4.0) using the pld gene sequence (NCBI GenBank accession number L16583). The forward primer sequence was AhF (5′-ATG TAC GAC GAT GAA GAC GCG-3′), the reverse sequence was AhR (5′-GCT TCC TTG TCG TTG AGA TTA TTA GC-3′), and the expected size of the amplicon of A. haemolyticum phospholipase D-specific PCR was 528 bp. The PCR mixture was used as described above. The PCR program was carried out as follows: one step of 10 min at 95°C; 30 cycles, with 1 cycle consisting of 30 s at 95°C, 60 s at 60°C, and 60 s at 72°C; and one step of 7 min at 72°C. All oligonucleotide primers used in this study were synthesized by MWG Biotech (Ebersberg, Germany).
RESULTS AND DISCUSSION
All seven bacterial cultures investigated in the present study could be identified phenotypically and genotypically as A. haemolyticum. Comparable to previous findings (18, 21), the seven strains produced a narrow zone of complete hemolysis on sheep blood agar plates, an enhanced hemolysis on rabbit blood agar plates, a synergistic CAMP-like hemolysis with Streptococcus agalactiae and Rhodococcus equi indicator strains and a reverse CAMP reaction with the beta-hemolysin of Staphylococcus aureus. In addition, the seven strains showed a moderate liquefaction of Loeffler agar which is well-known to be a typical property of A. pyogenes (2). However, the serum liquefaction caused by A. pyogenes DSM 20630 appeared to be enhanced, and A. haemolyticum DSM 20595 did not cause serum liquefaction. An extracellular protease of A. pyogenes had been isolated and further characterized by Schaufuss et al. (30). The extracellular substance causing the moderate serum liquefaction of the seven A. haemolyticum strains of the present study is not known. A cross-reaction with group G-specific antiserum could be observed, as already described (17), for the A. pyogenes control strain but not for the seven A. haemolyticum strains. The biochemical properties of the seven A. haemolyticum strains determined with the API Coryne test system are summarized in Table 2. The results corresponded to previous findings (10). The accuracy of the API Coryne test system for identification of A. haemolyticum has already been demonstrated (11, 12). The biochemical properties of the seven A. haemolyticum strains determined with the API Coryne test system could generally be confirmed with tablets containing substrates determining alkaline phosphatase, α-mannosidase, and pyrrolidonyl arylamidase enzyme activities and 4-methylumbelliferyl-conjugated substrates investigating the β-galactosidase, β-glucuronidase, and N-acetyl-β-glucosaminidase enzymes. In addition, the seven A. haemolyticum strains appeared to be positive for DNase and negative for catalase, hyaluronidase, and acetoin (Table 2). These tests confirmed the findings of Carlson and Kontiainen (3) and Carlson et al. (4) who recommended α-mannosidase diagnostic tablets and a negative Voges-Proskauer reaction for rapid identification of A. haemolyticum.
TABLE 2.
Biochemical properties of the seven A. haemolyticum strains and A. haemolyticum DSM 20595 investigated in the present study
| Biochemical property | Reactiona by:
|
|
|---|---|---|
| A. haemolyticum (n = 7) | A. haemolyticum DSM 20595 | |
| Nitrate reduction | −b | −b |
| Pyrazinamidase | (+) (4)b | +b |
| Pyrrolidonyl arylamidase | (+) (6)b; + (7)c | −b,c |
| Alkaline phosphatase | + (7)b,c | +b; −c |
| β-Glucuronidase | + (2)b,d | −b,d |
| β-Galactosidase | + (7)b,d | +b,d |
| α-Glucosidase | + (7)b | +b |
| N-Acetyl-β-glucosaminidase | + (7)b,d | +b,d |
| Esculin (β-glucosidase) | −b | −b |
| Urease | −b | −b |
| Gelatin | + (1)b, (+) (6)b | −b |
| Fermentation of: | ||
| Glucose | + (7)b | +b |
| Ribose | + (7)b | +b |
| Xylose | −b | −b |
| Mannitol | −b | −b |
| Maltose | + (7)b | +b |
| Lactose | + (7)b | +b |
| Saccharose | −b | −b |
| Glycogen | −b | −b |
| α-Mannosidase | + (7)c | +c |
| Catalase | − | − |
| Hyaluronidase | − | − |
| DNase | + (7) | + |
| Voges-Proskauer | − | − |
The reactions are shown as follows: +, positive reaction; (+), weak reaction; −, negative reaction. The number of positive strains is shown in parentheses after a positive reaction.
API Coryne test system (Biomerieux).
Tablets containing substrates (Inverness Medical).
4-Methylumbelliferyl-conjugated substrates (Sigma).
According to the results of the antibiotic resistance tests of the present study, all seven A. haemolyticum strains appeared to be susceptible to enrofloxacin, erythromycin, gentamicin, penicillin G, and tetracycline, and four strains appeared to be susceptible to sulfamethoxazole-trimethoprim. The resistance of two A. haemolyticum strains to sulfamethoxazole-trimethoprim and the susceptibility to most of the other antibiotics tested corresponded to the findings of others (5, 17, 35).
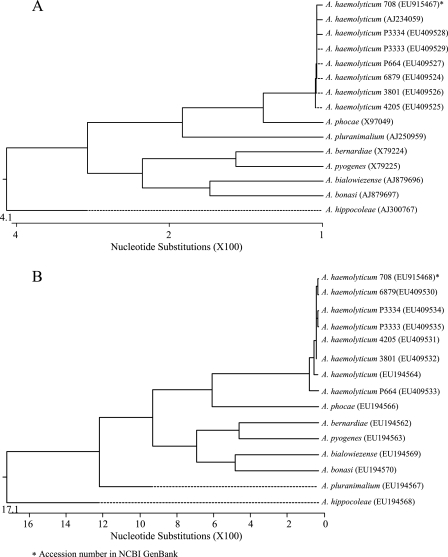
The seven A. haemolyticum strains could also be identified genotypically by amplification and sequencing of the 16S rRNA gene (NCBI GenBank accession numbers EU409524 to EU409529 and EU915467) and ISR (accession numbers EU409530 to EU409535 and EU915468) yielding an almost complete identity to the corresponding sequence of the A. haemolyticum reference strain obtained from GenBank. Dendrogram analysis of both sequencing results are shown in Fig. 1. Sequencing ISR of bacteria of genus Arcanobacterium and the design of ISR-specific oligonucleotide primers had already been used for PCR-mediated identification of A. bialowiezense and A. bonasi (13).
FIG. 1.
Dendrogram analysis of 16S rRNA gene sequences (A) and ISR sequences (B) of the seven A. haemolyticum strains of the present study and of eight Arcanobacterium reference strains obtained from NCBI GenBank.
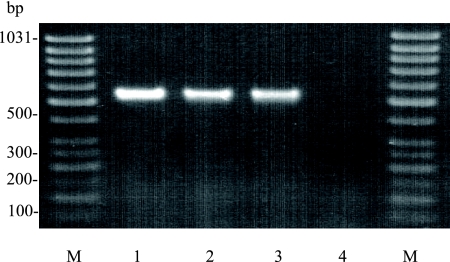
The species identification of the seven A. haemolyticum strains could also be confirmed by PCR-mediated amplification of species-specific regions of the pld gene encoding phospholipase D of A. haemolyticum. A typical amplicon (expected size of 528 bp) of the A. haemolyticum phospholipase D-specific PCR is shown in Fig. 2. No cross-reactivity could be observed with any of the other Arcanobacterium species or with both C. pseudotuberculosis reference strains (data not shown). A PCR-mediated identification of C. pseudotuberculosis using C. pseudotuberculosis pld-specific oligonucleotide primers had been described by Pacheco et al. (27). The gene encoding A. haemolyticum phospholipase D had been cloned and sequenced and showed some similarities to the corresponding genes of C. pseudotuberculosis and Corynebacterium ulcerans (9, 25). However, the A. haemolyticum phospholipase D gene-specific oligonucleotide primers designed in the present study could be used successfully for genotypic identification of this species and might improve a future diagnosis of A. haemolyticum infection in human and veterinary medicine.
FIG. 2.
Typical species-specific amplicons of the pld gene encoding A. haemolyticum phospholipase D with a size of approximately 530 bp (lanes 1, 2, and 3). A negative reaction was seen for A. pyogenes (lane 4). Lanes M, GeneRuler 50-bp DNA ladder (Fermentas).
To our knowledge, the present study is the first detailed phenotypic and genotypic characterization of A. haemolyticum strains isolated from infections of animals. However, at present, nothing is known about the route of infection and about the zoonotic potential these strains might have for the horse owner.
Footnotes
Published ahead of print on 19 November 2008.
REFERENCES
- 1.Barksdale, W. L., C. S. Cummins, and H. Harris. 1957. The mutation of Corynebacterium pyogenes to Corynebacterium haemolyticum. J. Gen. Microbiol. 16749-758. [DOI] [PubMed] [Google Scholar]
- 2.Bisping, W., and G. Amtsberg. 1988. Grampositive, sporenlose Stäbchen, p. 45-48. In W. Bisping and G. Amstberg (ed.), Farbatlas zur Diagnose bakterieller Infektionserreger der Tiere. Verlag Paul Parey, Berlin, Germany.
- 3.Carlson, P., and S. Kontiainen. 1994. Alpha-mannosidase: a rapid test for identification of Arcanobacterium haemolyticum. J. Clin. Microbiol. 32854-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlson, P., E. Eerola, and S. Kontiainen. 1995. Additional tests to differentiate Arcanobacterium haemolyticum and Actinomyces pyogenes. Zentralbl. Bakteriol. 282232-236. [DOI] [PubMed] [Google Scholar]
- 5.Carlson, P., S. Kontiainen, and O. V. Renkonen. 1994. Antimicrobial susceptibility of Arcanobacterium haemolyticum. Antimicrob. Agents Chemother. 38142-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlson, P., S. Kontiainen, O. V. Renkonen, A. Sivonen, and R. Visakorpi. 1995. Arcanobacterium haemolyticum and streptococcal pharyngitis in army conscripts. Scand. J. Infect. Dis. 2717-18. [DOI] [PubMed] [Google Scholar]
- 7.Chanter, N., N. Collin, N. Holmes, M. Binns, and J. Mumford. 1997. Characterization of the Lancefield group C streptococcus 16S-23S RNA gene intergenic spacer and its potential for identification and sub-specific typing. Epidemiol. Infect. 118125-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins, M. D., D. Jones, and G. M. Schofield. 1982. Reclassification of Corynebacterium haemolyticum (MacLean, Liebow & Rosenberg) in the genus Arcanobacterium gen. nov. as Arcanobacterium haemolyticum nom. rev., comb. nov. J. Gen. Microbiol. 1281279-1281. [DOI] [PubMed] [Google Scholar]
- 9.Cuevas, W. A., and J. G. Songer. 1993. Arcanobacterium haemolyticum phospholipase D is genetically and functionally similar to Corynebacterium pseudotuberculosis phospholipase D. Infect. Immun. 104310-4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding, H., and C. Lämmler. 1992. Evaluation of the API Coryne test system for identification of Actinomyces pyogenes. Zentralbl. Bakteriol. 39273-276. [DOI] [PubMed] [Google Scholar]
- 11.Freney, J., M. T. Duperron, C. Courtier, W. Hansen, F. Allard, J. M. Boeufgras, D. Monget, and J. Fleurette. 1991. Evaluation of API Coryne in comparison with conventional methods for identifying coryneform bacteria. J. Clin. Microbiol. 2938-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gavin, S. E., R. B. Leonard, A. M. Briselden, and M. B. Coyle. 1992. Evaluation of the rapid Coryne identification system for Corynebacterium species and other coryneforms. J. Clin. Microbiol. 301692-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hassan, A. A., H. Mohyla, T. Kanbar, J. Alber, C. Lämmler, A. Abdulmawjood, S. Speck, M. Zschöck, and R. Weiss. 2008. Molecular identification of Arcanobacterium bialowiezense and Arcanobacterium bonasi based on 16S-23S rRNA intergenic spacer region sequences. Vet. Microbiol. 130410-414. [DOI] [PubMed] [Google Scholar]
- 14.Hoyles, L., E. Falsen, G. Foster, F. Rogerson, and M. D. Collins. 2002. Arcanobacterium hippocoleae sp. nov., from the vagina of a horse. Int. J. Syst. Evol. Microbiol. 52617-619. [DOI] [PubMed] [Google Scholar]
- 15.Kostman, J. R., M. B. Alden, M. Mair, T. D. Edlind, J. J. LiPuma, and T. L. Stull. 1995. A universal approach to bacterial molecular epidemiology by polymerase chain reaction ribotyping. J. Infect. Dis. 171204-208. [DOI] [PubMed] [Google Scholar]
- 16.Kuhnert, P., S. E. Capaul, J. Nicolet, and J. Frey. 1996. Phylogenetic positions of Clostridium chauvoei and Clostridium septicum based on 16S rRNA gene sequences. Int. J. Syst. Bacteriol. 461174-1176. [DOI] [PubMed] [Google Scholar]
- 17.Lämmler, C., and H. Blobel. 1988. Comparative studies on Actinomyces pyogenes and Arcanobacterium haemolyticum. Med. Microbiol. Immunol. 177109-114. [DOI] [PubMed] [Google Scholar]
- 18.Lämmler, C., and H. Hartwigk. 1995. Actinomyces pyogenes und Arcanobacterium haemolyticum, p. 196-240. In H. Blobel and T. Schließer (ed.), Handbuch der bakteriellen Infektionen bei Tieren, vol. 2/3. Gustav Fischer Verlag, Stuttgart, Germany. [Google Scholar]
- 19.Lawson, P. A., E. Falsen, N. Weiss, and M. D. Collins. 2001. Arcanobacterium pluranimalium sp. nov., isolated from porpoise and deer. Int. J. Syst. Evol. Microbiol. 5155-59. [DOI] [PubMed] [Google Scholar]
- 20.Lehnen, A., H. J. Busse, K. Frölich, M. Krasinska, P. Kämpfer, and S. Speck. 2006. Arcanobacterium bialowiezense sp. nov. and Arcanobacterium bonasi sp. nov., isolated from the prepuce of European bison bulls (Bison bonasus) suffering from balanoposthitis, and emended description of the genus Arcanobacterium Collins et al. 1983. Int. J. Syst. Evol. Microbiol. 56861-866. [DOI] [PubMed] [Google Scholar]
- 21.Linder, R. 1997. Rhodococcus equi and Arcanobacterium haemolyticum: two “coryneform” bacteria increasingly recognized as agents of human infection. Emerg. Infect. Dis. 3145-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mackenzie, A., L. A. Fuite, F. T. H. Chan, J. King, U. Allen, N. MacDonald, and F. Diaz-Mitoma. 1995. Incidence and pathogenicity of Arcanobacterium haemolyticum during a 2-year study in Ottawa. Clin. Infect. Dis. 1177-181. [DOI] [PubMed] [Google Scholar]
- 23.MacLean, P. D., A. A. Liebow, and A. A. Rosenberg. 1946. A haemolytic Corynebacterium resembling Corynebacterium ovis and Corynebacterium pyogenes in man. J. Infect. Dis. 7969-90. [DOI] [PubMed] [Google Scholar]
- 24.Maddocks, J. L., and M. J. Greenan. 1975. A rapid method for identifying bacterial enzymes. J. Clin. Pathol. 28686-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McNamara, P. J., W. A. Cuevas, and J. G. Songer. 1994. Toxic phospholipases D of Corynebacterium pseudotuberculosis, C. ulcerans and Arcanobacterium haemolyticum: cloning and sequence homology. Gene 156113-118. [DOI] [PubMed] [Google Scholar]
- 26.NCCLS. 2004. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; information supplement. M31-S1. Vol. 24, no. 17. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 27.Pacheco, L. G. C., R. R. Pena, T. L. P. Castro, F. A. Dorella, R. C. Bahia, R. Caminati, M. N. L. Frota, S. C. Oliveira, R. Meyer, F. S. F. Alves, A. Miyoshi, and V. Azevedo. 2007. Multiplex PCR assay for identification of Corynebacterium pseudotuberculosis from pure cultures and for rapid detection of this pathogen in clinical samples. J. Med. Microbiol. 56480-486. [DOI] [PubMed] [Google Scholar]
- 28.Ramos, C. P., G. Foster, and M. D. Collins. 1997. Phylogenetic analysis of the genus Actinomyces based on 16S rRNA gene sequences: description of Arcanobacterium phocae sp. nov., Arcanobacterium bernardiae comb. nov., and Arcanobacterium pyogenes comb. nov. Int. J. Syst. Bacteriol. 4746-53. [DOI] [PubMed] [Google Scholar]
- 29.Raus, J., and D. N. Love. 1983. Characterization of coagulase-positive Staphylococcus intermedius and Staphylococcus aureus isolated from veterinary clinical specimens. J. Clin. Microbiol. 18789-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schaufuss, P., R. Sting, and C. Lämmler. 1989. Isolation and characterization of an extracellular protease of Actinomyces pyogenes. Zentralbl. Bakteriol. 4452-459. [DOI] [PubMed] [Google Scholar]
- 31.Skov, R. L., A. K. Sanden, V. H. Danchell, K. Robertsen, and T. Ejlertsen. 1998. Systemic and deep-seated infections caused by Arcanobacterium haemolyticum. Eur. J. Clin. Microbiol. Infect. Dis. 17578-582. [DOI] [PubMed] [Google Scholar]
- 32.Slifkin, M., and G. M. Gil. 1983. Rapid biochemical tests for the identification of groups A, B, C, F, and G streptococci from throat cultures. J. Clin. Microbiol. 1829-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan, T. Y., S. Y. Ng, H. Thomas, and B. K. Chan. 2006. Arcanobacterium haemolyticum bacteraemia and soft-tissue infections: case report and review of the literature. J. Infect. 5369-74. [DOI] [PubMed] [Google Scholar]
- 34.Tyrrell, K. L., D. M. Citron, J. R. Jenkins, and E. J. Goldstein. 2002. Periodontal bacteria in rabbit mandibular and maxillary abscesses. J. Clin. Microbiol. 401044-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vargas, J., M. Hernandez, C. Silvestri, O. Jiménez, N. Guevara, M. Carballo, N. Rojas, J. Riera, E. Alayo, M. Fernández, A. J. Rodriguez-Morales, and M. Silva. 2006. Brain abscess due to Arcanobacterium haemolyticum after dental extraction. Clin. Infect. Dis. 121810-1811. [DOI] [PubMed] [Google Scholar]