Figure 1.
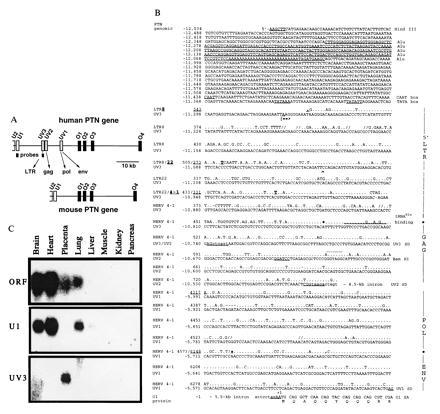
Insertion of HERV into the human PTN gene and tissue-specific distribution of HERV-PTN fusion transcripts. (A) Comparison of the genomic organization of the human and murine PTN genes. Homologous exons corresponding to the ORF (O1 to O4) or 5′-UTR (U2, U1) of the murine (24) and the human gene (20, 21), as well as the HERV-derived exons (UV3, UV2, and UV1) and the position of exon-specific probes, are shown. (B) Sequence comparison between the HERV insert in the human PTN gene and type C HERV fragments LTR8, LTR22, and HERV4-1 (22, 23) using the GCG program. Gaps (//) were introduced for optimal alignment. The PTN gene numbering system (GenBank accession nos. U71455U71455 and U71456U71456) on the left refers to the first nucleotide of the translation start codon as +1. The viral sequence positions were taken from the respective GenBank entries for LTR8, LTR22, and HERV4-1 (accession nos. M32219M32219, M32220M32220, and K02168K02168, respectively). A 280-nt Alu sequence in the promoter region and CAAT and TATA boxes are underlined and intronic sequence is denoted by lowercase type. The transcription start site (|==>) was mapped by primer extension. Similarity to retroviral elements is indicated on the right and the respective nucleotide positions separating the different elements are highlighted in the sequence. The tRNAGlu primer binding site, UV3 cDNA end (∧), restriction sites used for generation of the promoter-reporter constructs, and the N-terminal sequence of the PTN protein are shown. The splice donor (SD) sites of UV3, UV2, and UV1, as well as the splice acceptor (SA) in exon O1, are underlined and the retroviral SD of HERV 4–1 is indicated in addition (by dots above the sequence). (C) Northern blot analysis of mRNA from various human tissues using exon-specific probes.
