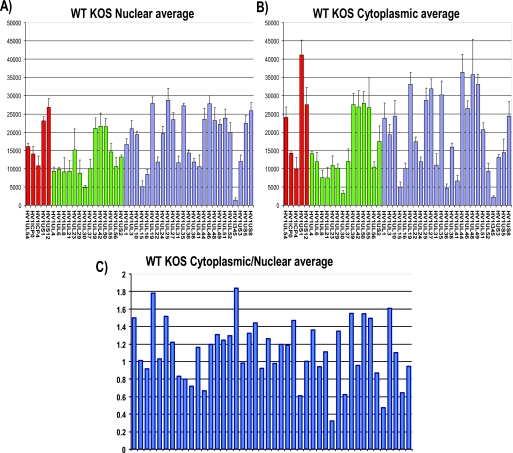
FIG. 7.
Microarray analysis of the cytoplasmic/nuclear ratio for HSV-1 transcripts. (A, B) HeLa cells were infected with wild-type (WT) HSV-1 KOS for 8 h. Cells were lysed, and nuclear and cytoplasmic fractions were prepared as described previously (30). RNA was isolated from each fraction, and poly(A)+ RNA was selected and reverse transcribed as described previously (38). The cDNA from each fraction was hybridized to HSV-1-specific microarray chips as described previously (38) and quantified by using Array Vision software. Experiments were performed in triplicate, and the average expression value for each transcript from each fraction was calculated and plotted, with the y axis representing the intensity of the light scattering signal and the x axis representing individual HSV-1 transcripts. Error bars show standard deviations. Red bars represent immediate early transcripts, green bars show early transcripts, and blue bars show late transcripts. (C) The cytoplasmic/nuclear ratio for each transcript was calculated from the average values for each transcript from the cytoplasmic and nuclear fractions. The ratio for each transcript was plotted on the y axis, with 1.0 indicating equal abundance in the cytoplasm and nucleus. A ratio greater than 1.0 indicates that a transcript was more abundant in the cytoplasm than in the nucleus. The x axis represents individual transcripts in the same order as in panels A and B.