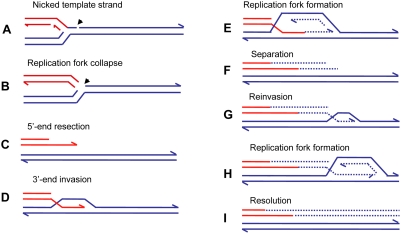
Figure 4. Repair of a collapsed replication fork by BIR.
When a replication fork encounters a nick in a template strand (A) (arrowhead), one arm of the fork breaks off (red), producing a collapsed fork (B). At the single double-strand end, the 5′ strand is resected, giving a 3′ overhang (C). The 3′ single-strand end invades the sister molecule (blue), forming a D-loop (D), which subsequently becomes a replication fork with both leading and lagging strand replication (E). There is a Holliday junction at the site of the D-loop. Migration of the Holliday junction, or some other helicase activity, separates the extended double-strand end from its templates (F). The separated end is again processed to give a 3′ single-strand end, which again invades the sister, and forms a replication fork (G). Eventually the replication fork becomes fully processive, and continues replication to the chromosome end (H and I). This process is shown here with the Holliday junction following the fork so that newly formed strands are segregated together (conservative segregation) (H). Each line represents a DNA nucleotide chain (strand). Polarity is indicated by half arrows on 3′ end. New DNA synthesis is shown by dashed lines. The publications on which this model is based are cited in the text.