Figure 2.
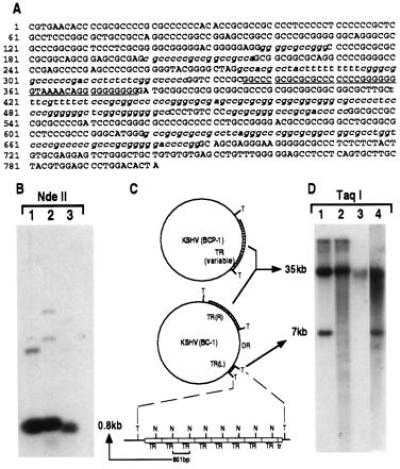
(A) Sequence of TR unit demonstrating its high G+C content. Sequences highly homologous to conserved herpesvirus pac1 sites are underlined, with lesser homology sites to specific pac1 and pac2 sequences italicized. (B) Southern blot of DNA from BC-1 (lane 1), BCP-1 (lane 2) and a KS lesion (lane 3) digested with NdeII which cuts once in the TR sequence and probed with a plasmid containing the TR sequence. The intense hybridization band at 0.8 kb represents multiple copies of the NdeII-digested single unit TR (C). A schematic representation (C) of genome structures of KSHV in BCP-1 and BC-1 cell lines consistent with the data presented in B and D. Taq I (T) sites flank the TR regions and NdeII (N) sites are within the TRs. Lowercase tr refers to the deleted truncated TR unit at the left end of the unique region. DR represents the duplicated region of the LUR buried within the TR. (D) Southern blot hybridization with TR probe of DNA from BC-1 (lane 1), BCP-1 (lane 2), a KS lesion (lane 3), and HBL-6 (lane 4) digested with Taq I, which does not cut in the TR. Taq I-digested DNA from both BC-1 (lane 1) and HBL-6 (lane 4) show similar TR hybridization patterns, suggesting identical insertion of a unique sequence into the TR region, which sequencing studies demonstrate is a duplicated portion of the LUR (see text). BCP-1 TR hybridization (lane 2) shows laddering consistent with a virus population having variable TR region lengths within this cell line due to lytic replication. The absence of TR laddering in KS lesion DNA (lane 3) suggests that a clonal virus population is present in the tumor.
