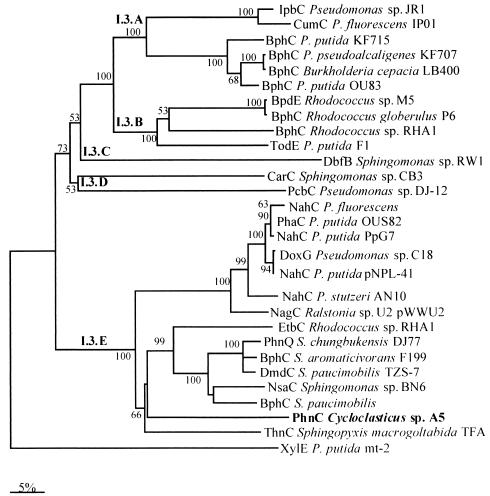
FIG. 3.
Dendrogram based on the extradiol dioxygenases. The dendrogram was constructed by using the previously published amino acid sequences processed by Clustal W. Scale bar = 5% divergence. The numbers at nodes are bootstrap values expressed as percentages. The following sequences were used: Pseudomonas sp. strain JR1 ipbC gene (GenBank accession no. U53507), Pseudomonas fluorescens IP01 cumC gene (D37828), P. putida KF715 bphC gene (M33813), Pseudomonas pseudoalcaligenes KF707 bphC gene (M83673), Burkholderia cepacia LB400 bphC gene (X66122), P. putida OU83 bphC gene (X91876), Rhodococcus sp. strain M5 bpdE gene (U27591), Rhodococcus globerulus P6 bphC gene (X75663), Rhodococcus sp. strain RHA1 bphC gene (D32142), P. putida F1 todE gene (JO4996), Sphingomonas sp. strain RW1 dbfB gene (X72850), Sphingomonas sp. strain CB3 carC gene (AF060489), Pseudomonas sp. strain DJ-12 pcbC gene (D44550), P. fluorescens nahC gene (AY048760), P. putida OUS82 pahC gene (D16629), P. putida PpG7 nahC gene (J04994), Pseudomonas sp. strain C18 doxG gene (M60405), P. putida pNPL41 nahC gene (Y14173), P. stutzeri AN10 nahC gene (AF039533), Ralstonia sp. strain U2 nagC gene (AF036940), Rhodococcus sp. strain RHA1 etbC gene (D78322), Sphingomonas chungbukensis DJ77 phnQ gene (AF061802), S. aromaticivorans F199 bphC gene (AF079317), S. paucimobilis TZS-7 dmdC gene (AB035677), Sphingomonas sp. strain BN6 nsaC gene (U65001), S. paucimobilis bphC gene (P11122), S. macrogoltabida TFA thnC gene (AF157565), and P. putida mt-2 xylE gene (V01161).