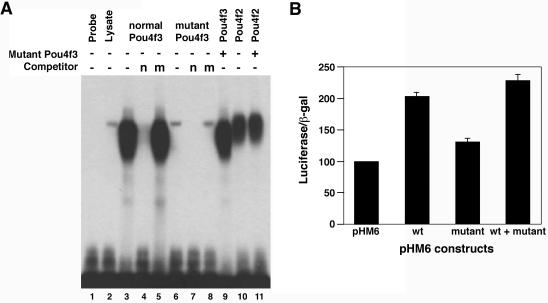
FIG. 2.
DNA binding and transcriptional activities of wild-type and mutant POU4F3 proteins. (A) EMSA was performed with a radiolabeled POU4F binding site as a probe. Reactions contained in vitro-translated normal POU4F3 (lanes 3 to 5) or mutant POU4F3 (lanes 6 to 8). Competition was performed by using a 500-fold excess amount of cold consensus POU4F binding site (n; lanes 4 and 7) or nonspecific competitor (m; lanes 5 and 8). To assess whether mutant POU4F3 interferes with DNA binding, reactions containing in vitro translated POU4F3 (lane 3 and 9) and POU4F2 (lanes 10 and 11) were incubated in the presence (lanes 9 and 11) or absence (lanes 3 and 10) of in vitro-translated mutant POU4F3. (B) The transcriptional activity is shown as relative luciferase activity after cotransfection of different expression constructs with the pGL2-SNAP25 reporter into PC12 cells. Luciferase activity was measured 48 h after transfection. The transfection efficiency was controlled by measuring β-galactosidase activity after transfection with the pHM6-lacZ plasmid. The basic transcriptional activity for the promoter with pHM6 empty vector was set arbitrarily to 100. The data shown represent three separate experiments performed in triplicate in each experiment (mean ± the standard deviation [SD]).